TAG Protein Degradation - aTAG, dTAG, BromoTAG®
Our TAG Degradation technologies (dTAG, aTAG and BromoTag®) offer a generalizable strategy to degrade, in principle, any intracellular protein of interest. The key benefit is that these techniques do not rely on the pre-existence of a ligand or PROTAC® for the protein of interest. The TAG Degradation Platform is a useful alternative to genetic knockdown/knockout methods for target validation and can be used in cell culture or in vivo. The most recent addition to our TAG Degraders product line is BromoTag, exclusively licensed from University of Dundee.
Why Use TAG Degraders?
dTAG, aTAG and BromoTag Degraders can be used for target validation and exploration, offering an attractive alternative to genetic knockdown/knockout. The table below provides a comparison of TAG Degradation with commonly used genetic methods, including CRISPR/Cas9 and RNA interference. Key advantages of TAG Degraders include the ability to tune the extent of protein knockdown by varying the dose, and the more rapid onset of action (kinetics) for studying ‘fast biology’. In addition, dTAG, aTAG and BromoTag Degraders can be washed out of cell culture media, reversing their effect. TAG Degraders can, in theory, be used orthogonally to knock down more than one target protein within a cell.
A Comparison of TAG Degradation with Genetic Methods of Protein Knockdown
Dose Tuneability | Efficacy | Reversibility | Kinetics | Selectivity | |
|---|---|---|---|---|---|
| TAG Degradation Platforms (dTAG/aTAG/BromoTag) | *** | **** | **** | *** | **** |
| Gene Knockout e.g. CRISPR/Cas | * | **** | * | * | **** |
| Gene Knockdown e.g. RNAi | * | *** | * | * | **** |
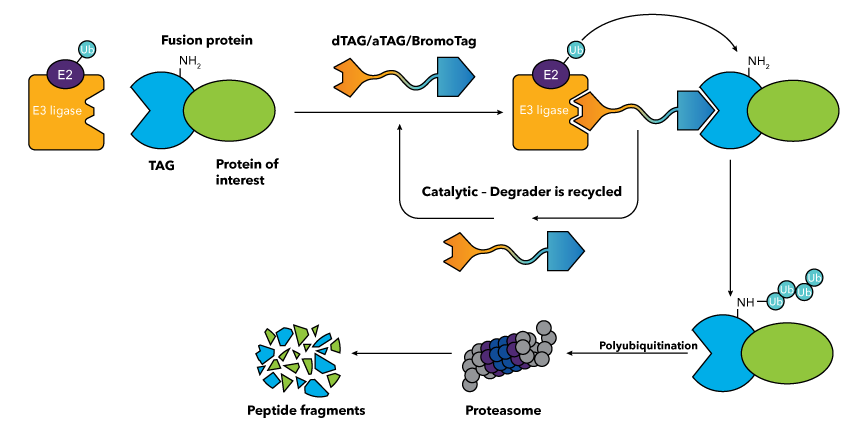
How do dTAG, aTAG and BromoTag Degraders Work?
The different TAG Degradation platforms all require the protein of interest to be expressed as a fusion with a TAG protein (also known as a degron tag) via transgene expression or CRISPR-mediated locus-specific knock-in (see resources section below for protocols). Subsequent treatment with the relevant TAG Degrader targets the entire fusion protein for degradation. Heterobifunctional TAG Degraders mediate the formation of a ternary complex between an E3 ubiquitin ligase, such as cereblon (CRBN) or von Hippel Lindau (VHL) protein, and the fusion protein. This leads to polyubiquitination of the target protein and the subsequent degradation of the entire fusion protein by the proteasome. dTAG, aTAG and BromoTag Degraders act catalytically, repeatedly engaging and directing the ubiquitination of target molecules. They are cell-permeable and are suitable for in vitro and in vivo applications. The key difference between the dTAG, aTAG and BromoTag technologies is the identity of the TAG protein used.
dTAG
The dTAG (or degradation TAG) system utilizes a single-point mutant of (F36V) as the TAG domain with corresponding Degraders that selectively recruit FKBP12F36V over wild-type FKBP12. dTAG Degraders recruiting two different E3 ligases are available: dTAG-13 (Catalog # 6605) and dTAG-7 (Catalog # 6912) recruit CRBN while dTAGV-1 (Catalog # 6914) recruits VHL. Negative controls are available.
aTAG
The aTAG (Achilles TAG) system, uses the enzyme MTH1 (MutT homolog-1; NUDT1) as the degron TAG, which is expressed as a fusion with the protein of interest. MTH1 is used as the TAG in this system since the loss of this protein has no known phenotypic consequence. aTAG Degraders, aTAG 2139 (Catalog # 6970) and aTAG 4531 (Catalog # 6971) are heterobifunctional molecules comprising a ligand selective for MTH1 linked to a CRBN (E3 ligase) ligand and bring about potent and rapid degradation of the fusion protein by the proteasome.
BromoTag
Within the BromoTag system the degron tag is a modified BET bromodomain, Brd4BD2 L387A. The Degrader BromoTag® AGB1 (Catalog # 7686) recruits the modified Brd4BD2 L387A-target protein fusion for ubiquitination by VHL E3 ligases, leading to its rapid proteasomal degradation. BromoTag® AGB1 is highly selective for Brd4BD2 L387A over wild-type Brd4. The BromoTag technology is made available by Bio-Techne through an exclusive agreement with the University of Dundee (UK).
TAG Degradation Products Available from Bio-Techne
| Degraders | |||
|---|---|---|---|
| 7893 | 5-Ph-IAA-AM Selective and potent TAG Degrader for auxin-inducible degron 2 system; 5-Ph-IAA analog | 7530 | dTAG-47 Degrades mutant FKBP12F36V fusion proteins; useful alternative to genetic methods for target validation |
| 6970 | aTAG 2139 Degrader of MTH1 fusion proteins for use within the aTAG system | 6912 | dTAG-7 First generation Degrader for mutant FKBP12F36V fusion proteins; useful alternative to genetic methods for target validation |
| 6971 | aTAG 4531 Degrader of MTH1 fusion proteins for use within the aTAG system | 6914 | dTAGV-1 Potent and selective degrader of mutant FKBP12F36V fusion proteins |
| 7686 | BromoTag® AGB1 Potent and selective Degrader of Brd4BD2 L387A fusion proteins (BromoTag®) | 7374 | dTAGV-1 hydrochloride Hydrochloride salt of dTAGV-1 (Catalog # 6914); suitable for in vivo use |
| 7688 | BromoTag® AGB3 Selective Degrader of Brd4BD2 L387A fusion proteins (BromoTag®) | 7392 | 5-Ph-IAA Selective and potent TAG Degrader for auxin-inducible degron 2 system |
| 6605 | dTAG-13 Degrades mutant FKBP12F36V fusion proteins; useful alternative to genetic methods for target validation | 7669 | XY-06-007 Potent and selective Degrader of Brd4BD1 L94V fusion proteins |
| Controls | |||
| 7575 | aTAG 2139-NEG Negative control for aTAG 2139 (Catalog # 6970) | 7687 | BromoTag® cis-AGB1 Negative control for BromoTag® AGB1 (Catalog # 7686) |
| 7687 | BromoTag® cis-AGB1 Negative control for BromoTag® AGB1 (Catalog # 7686) | 6916 | dTAG-13-NEG Negative control for dTAG-13 (Catalog # 6605) |
| 6916 | dTAG-13-NEG Negative control for dTAG-13 (Catalog # 6605) | ||
| Other | |||
| 6207 | AP 1867 Selective binding ligand for FKBP12F36V | 7883 | dTAG-Biotin Affinity probe for mutant FKBP12F36V proteins |
| 8102 | dTAG Janelia Fluor® 525 Fluorogenic srTAG probe for live cell imaging of FBKP12F36V/L labeled proteins | 7892 | dTAG-Fluorescein Fluorescent probe for labeling mutant FKBP12F36V proteins |
| 8103 | dTAG Janelia Fluor® 585 Fluorogenic srTAG probe for live cell imaging of FBKP12F36V/L labeled proteins | 7787 | Ortho AP 1867 Selective binding ligand for FKBP12F36V; precursor for dTAG compounds |
| 8101 | dTAG Janelia Fluor® 635 Fluorogenic srTAG probe for live cell imaging of FBKP12F36V/L labeled proteins | ||
PROTAC® is a registered trademark of Arvinas Operations, Inc., and is used under license.
BromoTag® is a registered trademark of the University of Dundee, and is used under license.
Janelia Fluor® is a registered trademark of Howard Hughes Medical Institute.
Related Resources
Validating Targets for Targeted Protein Degradation using dTAG Whitepaper
Peer Reviewed Research Paper: Development of an AchillesTAG degradation system and its application to control CAR-T activity
Visit Bio-Techne brand Tocris to view the protocol for the CRISPR-mediated specific knock-in of MTH1-fusion proteins
Visit Addgene.org to find plasmid vectors for the lentiviral expression and CRISPR-mediated knock-in of FKBP12F36V
-
Bensimon, A et al. (2020) Targeted degradation of SLC transporters reveals amenability of multi-pass transmembrane proteins to ligand-induced proteolysis. Cell Chem.Biol. PMID: 32386596.
-
Bond et al. (2021) Development of BromoTag: a 'Bump-and-Hole'-PROTAC system to induce potent, rapid, and selective degradation of tagged target proteins. J. Med. Chem. PMID: 34652918.
-
Nabet, B et al. (2020) Rapid and direct control of target protein levels with VHL-recruiting dTAG molecules. Nat. Commun. PMID: 32948771.
-
Nabet, B et al. (2018) The dTAG system for immediate and target-specific protein degradation. Nat. Chem. Biol. PMID: 29581585.