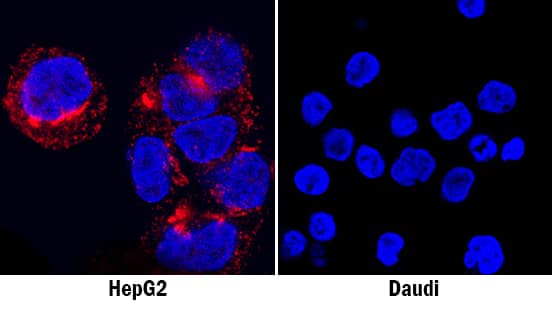
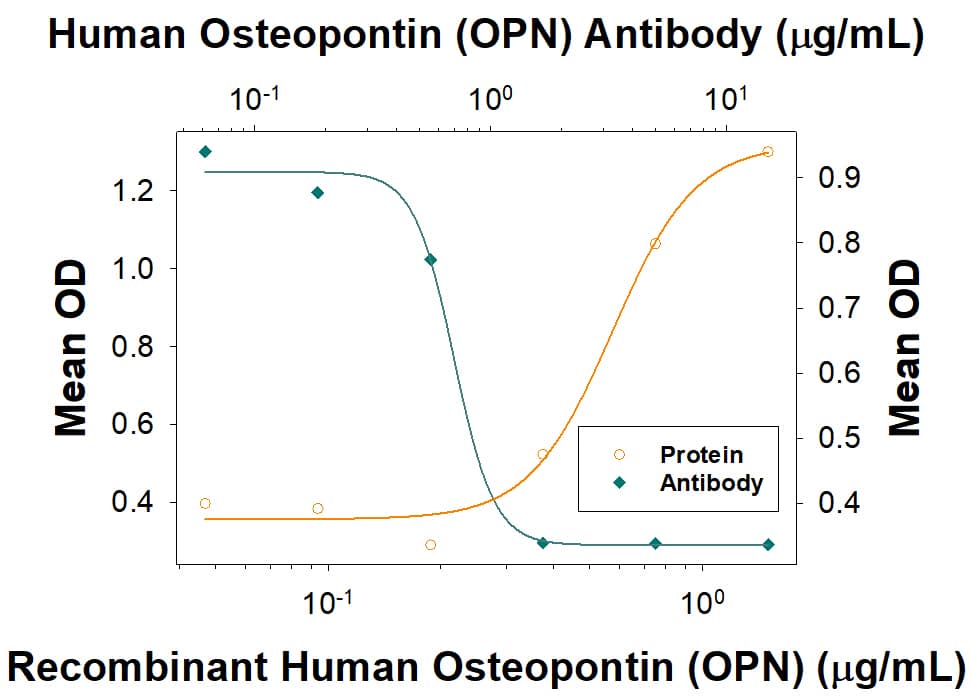
Cell Adhesion Mediated by Osteopontin/OPN and Neutralization by Human Osteopontin/OPN Antibody.
Recombinant Human Osteopontin/OPN (Catalog #
1433-OP), immobilized onto a microplate, supports the adhesion of the HEK293 human embryonic kidney cell line in a dose-dependent manner (orange line). Adhesion elicited by Recombinant Human Osteopontin/OPN (1 µg/mL) is neutralized (green line) by increasing concentrations of Goat Anti-Human Osteopontin/OPN Antigen Affinity-purified Polyclonal Antibody (Catalog # AF1433). The ND
50 is typically 0.300 - 3.60 µg/mL.
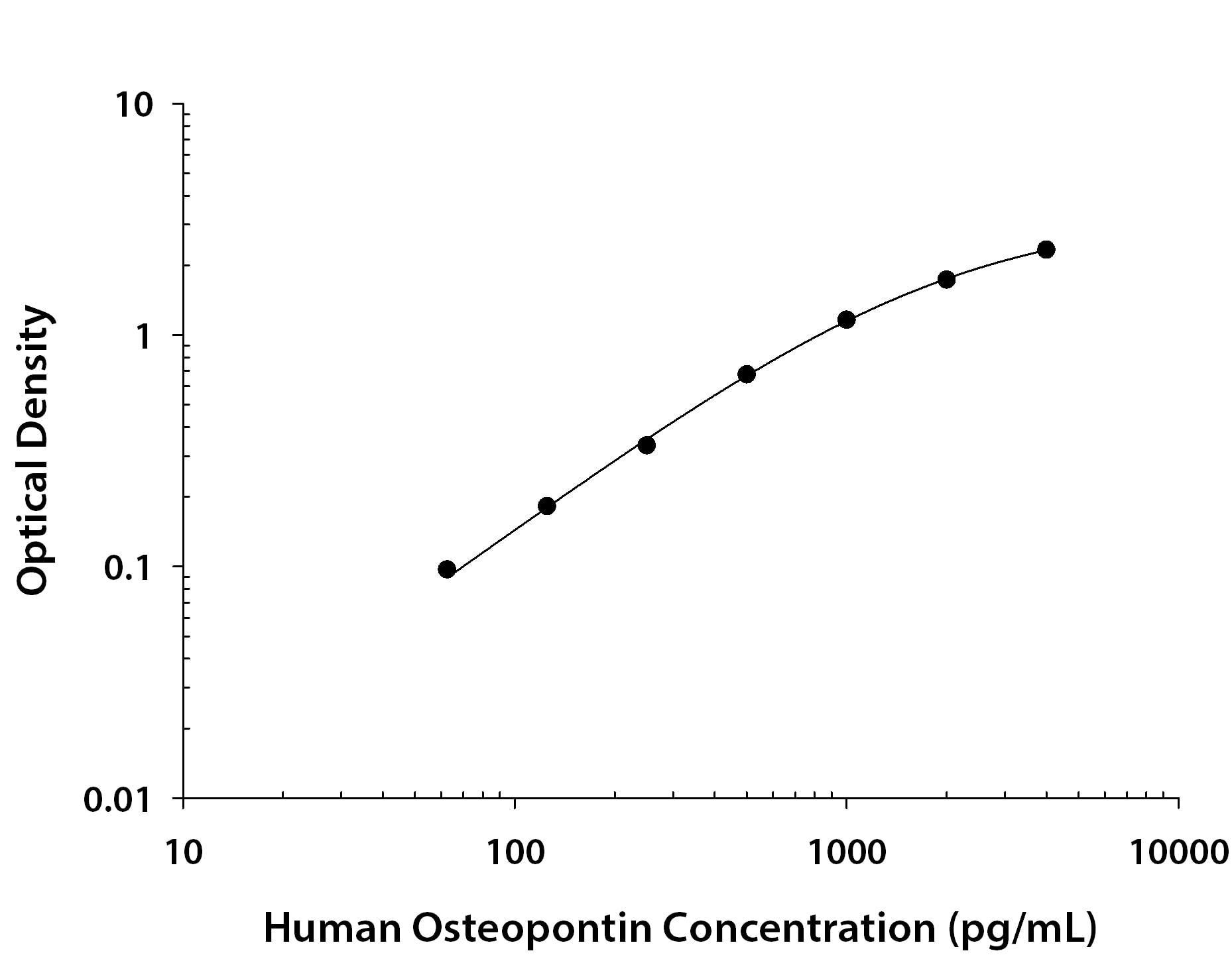
Human Osteopontin/OPN ELISA Standard Curve.
Recombinant Human Osteopontin/OPN protein was serially diluted 2-fold and captured by Mouse Anti-Human Osteopontin/OPN Monoclonal Antibody (Catalog #
MAB14332R) coated on a Clear Polystyrene Microplate (Catalog #
DY990). Goat Anti-Human Osteopontin/OPN Antigen Affinity-purified Polyclonal Antibody (Catalog # AF1433) was biotinylated and incubated with the protein captured on the plate. Detection of the standard curve was achieved by incubating Streptavidin-HRP (Catalog #
DY998) followed by Substrate Solution (Catalog #
DY999) and stopping the enzymatic reaction with Stop Solution (Catalog #
DY994).
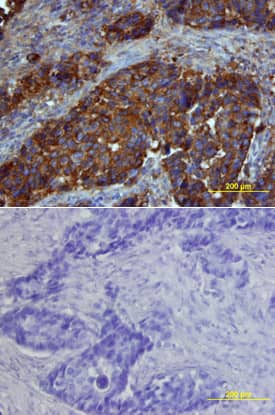
Detection of Osteopontin/OPN in Human Kidney.
Formalin-fixed paraffin-embedded tissue sections of human kidney were probed for OPN mRNA (ACD RNAScope Probe, catalog #420101; Fast Red chromogen, ACD catalog # 322750). Adjacent tissue section was processed for immunohistochemistry using goat anti-human OPN polyclonal antibody (R&D Systems catalog #
AF1433) at 1ug/mL with overnight incubation at 4 degrees Celsius followed by incubation with anti-goat IgG VisUCyte HRP Polymer Antibody (Catalog #
VC004) and DAB chromogen (yellow-brown). Tissue was counterstained with hematoxylin (blue). Specific staining was localized to cytoplasm in tubules.
Detection of Human Osteopontin/OPN by Western Blot
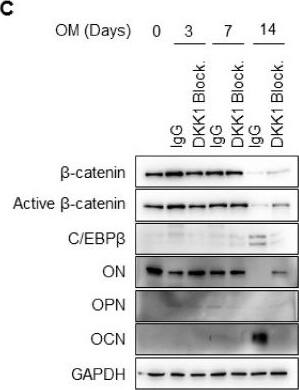
DKK1 blockade inhibits mineralization of osteoblast differentiation. Osteoblasts were treated with 1 μg anti-DKK1 or IgG as controls during osteoblast differentiation (n = 3). At indicated days, analysis of (A) results of ALP and ARS staining; scale bar is 200 μm, (B) hydroxyapatite staining; BF, Bright Field; HA, Hydroxyapatite; Scale bar is 500 μm, (C) immunoblotting for proteins, and (D) qPCR for mRNA. Representative data are shown (n = 3). * p < 0.05 (mean ± SD; n = 3). Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/31963554), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Osteopontin/OPN by Western Blot
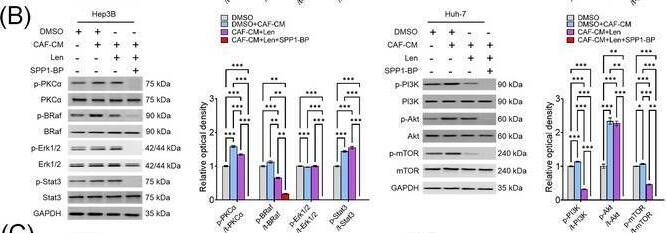
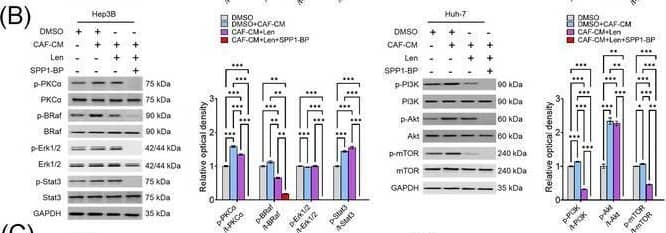
Molecular mechanism underlying CAF‐derived SPP1‐induced resistance to sorafenib or lenvatinib. (A‐B) Changes in the expression levels of PKC alpha, BRAF/ERK/STAT3, and PI3K/AKT/mTOR pathway proteins in CAF‐CM‐incubated Huh‐7 cells treated with sorafenib (15 μmol/L; A) or lenvatinib alone (5 μmol/L; B) and in combination with SPP1‐BP. (C‐D) Changes in the phosphorylation of AKT, mTOR, BRAF, and ERK1/2 under ITGB1 and/or ITGB5 silencing in Hep3B and Huh‐7 cells treated with sorafenib (C) or lenvatinib (D) in combination with CAF‐CM. (Mean ± SEM; Two‐way ANOVA test; *P < 0.05; **P < 0.01; ***P < 0.001).Abbreviations: DMSO, dimethyl sulfoxide; CAF, cancer‐associated fibroblast; CM, culture medium; Sor, sorafenib; SPP1‐BP, SPP1‐blocking peptide, PKC alpha, protein kinase C alpha; BRAF,v‐Raf murine sarcoma viral oncogene homolog B; ERK, extracellular signal‐related kinase; STAT3, signal transducer and activator of transcription 3; GAPDH, glyceraldehyde 3‐phosphate dehydrogenase; PI3K, phosphatidylinositol‐3‐kinase; AKT, protein kinase B; mTOR, mammalian target of rapamycin; Len, lenvatinib; siCtrl, negative control; ITGB1, integrin subunit beta1; ITGB5, integrin subunit beta5. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/36919193), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Osteopontin/OPN by Immunohistochemistry
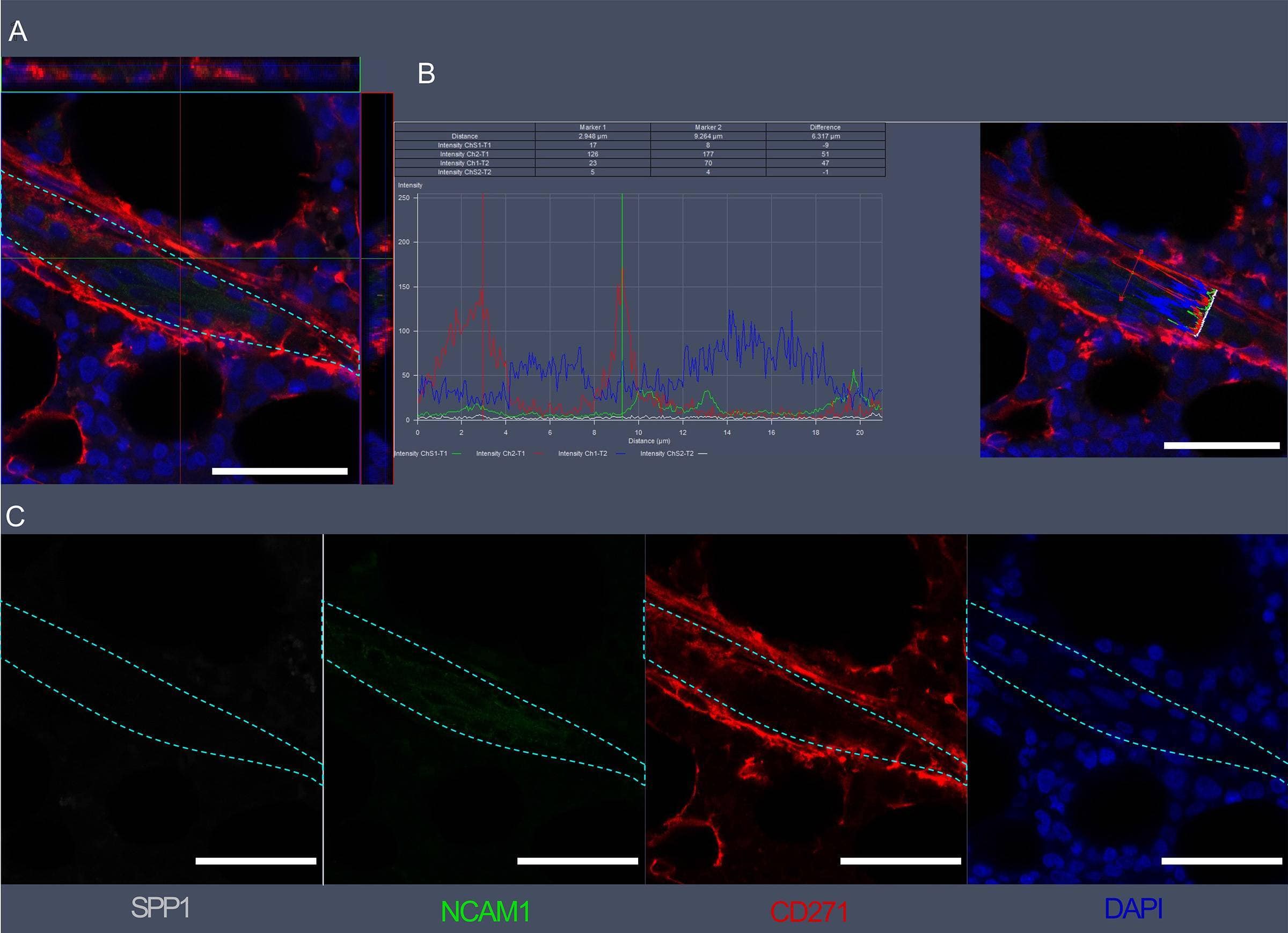
Visualization of SPP1, NCAM1, and CD271 expression in bone marrow vascular regions by confocal microscopy.(A) Confocal scan of vascular region in BM biopsies with 3D orthographic cross-section view, co-stained with rabbit anti-NCAM1, mouse anti-CD271, goat anti-SPP1, and DAPI. (B) Intensity profile for all channels in A across a cell of interest in a representative z plane. (C) Single channel data for the florescent markers in A. Scale bars represent 50 μm. Cyan dashed lines indicate vessel surface. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/36876630), licensed under a CC-BY license. Not internally tested by R&D Systems.
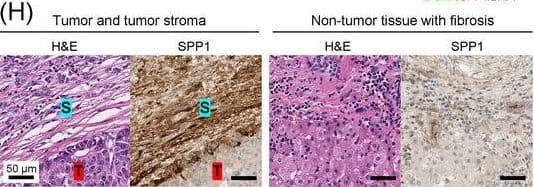
Detection of Osteopontin/OPN by Immunohistochemistry
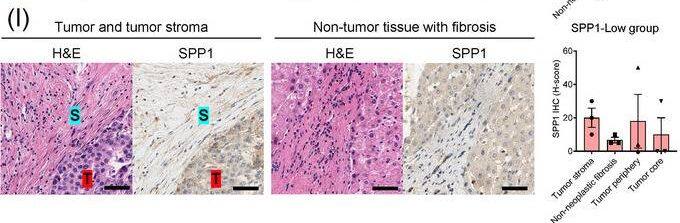
Identification of the CAF‐derived molecules that induce resistance to sorafenib or lenvatinib in patients with HCC. (A) Heat map of 790 genes overexpressed in CAFs. (B) GSEA of the genes overexpressed in CAFs. (C) Process of candidate gene selection. (D) Expression levels of the 9 candidate genes according to sorafenib response in the GSE109211 dataset. (E) Comparison of SPP1 expression between sorafenib responders and non‐responders in the GSE109211 and GSE143233 datasets. (F) Immunofluorescence staining of alpha‐SMA and SPP1 in tumor tissues from patients with HCC. (G) Comparison of SPP1 expression between CAFs and their paired para‐cancer fibroblasts in the WTS data from the 9 pairs of CAF and para‐cancer fibroblasts. (H) Representative H&E and SPP1 IHC images of the tumor (T), tumor stroma (S) (left), and non‐tumor fibrous tissue (middle) in an SPP1‐high patient. Scale bar, 50 μm. Comparison of SPP1 IHC staining intensity according to tumor location in the SPP1‐high group (right). (I) Representative H&E and SPP1 IHC images of tumor (T), tumor stroma (S) (left), and non‐tumor fibrous tissue (middle) in an SPP1‐low patient. Comparison of SPP1 IHC staining intensity according to tumor location in the SPP1‐low group (right). (Mean ± SEM; unpaired Welch's t‐test; *P < 0.05; **P < 0.01; ***P < 0.001).Abbreviations: HCC, hepatocellular carcinoma; NF, normal fibroblast; CAFs, cancer‐associated fibroblasts; ELISA, enzyme linked immunosorbent assay; H&E, hematoxylin and eosin; alpha‐SMA, alpha smooth muscle actin; SPP1, secreted phosphoprotein 1; DAPI, 4′, 6‐diamidino‐2‐phenylindole; PAF, para‐cancer fibroblast; S, tumor stroma; T, tumor; IHC, immunohistochemistry. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/36919193), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Osteopontin/OPN by Western Blot
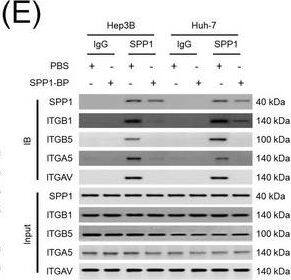
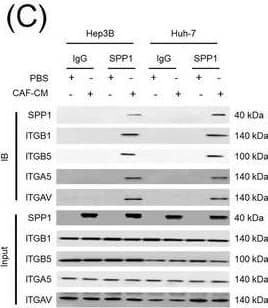
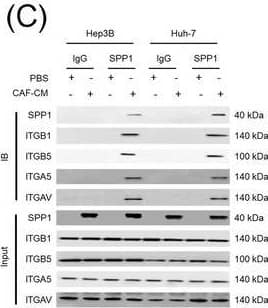
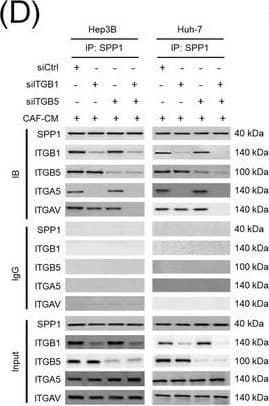
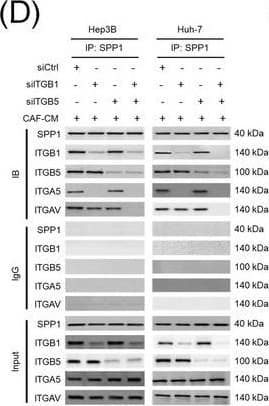
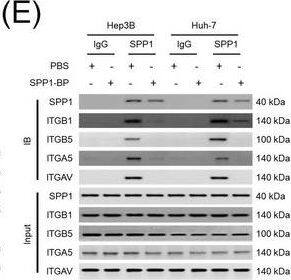
Integrin complexes (integrin alphaV beta5, alpha5 beta1, and alphaV beta1) on HCC cells were identified as binding proteins of SPP1. (A) Western blotting of SPP1 receptor‐binding protein expression in HCC cells. (B) Measurement of CD44 and integrin expression in Hep3B and Huh‐7 cells by fluorescent intensity. (C‐E) Co‐IP of SPP1 after CAF‐CM treatment, followed by Western blotting for SPP1, ITGA5, ITGAV, ITGB1, and ITGB5 in HCC cells (C), HCC cells silenced for ITGB1 and/or ITGB5 (D), and HCC cells incubated with/without SPP1‐BP (E).Abbreviations: ITGA4, integrin subunit alpha4; ITGA5, integrin subunit alpha5; ITGA8, integrin subunit alpha8; ITGA9, integrin subunit alpha9; ITGAV, integrin subunit alphaV; ITGB1, integrin subunit beta1; ITGB3, integrin subunit beta3; ITGB5, integrin subunit beta; GAPDH, glyceraldehyde 3‐phosphate dehydrogenase; RFU, relative fluorescence units; IgG, immunoglobulin G; SPP1, secreted phosphoprotein 1; CAF, cancer‐associated fibroblast; CM, culture medium; IB, immunoblotting; siCtrl, negative control; SPP1‐BP, SPP1‐blocking peptide. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/36919193), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Osteopontin/OPN by Western Blot
Integrin complexes (integrin alphaV beta5, alpha5 beta1, and alphaV beta1) on HCC cells were identified as binding proteins of SPP1. (A) Western blotting of SPP1 receptor‐binding protein expression in HCC cells. (B) Measurement of CD44 and integrin expression in Hep3B and Huh‐7 cells by fluorescent intensity. (C‐E) Co‐IP of SPP1 after CAF‐CM treatment, followed by Western blotting for SPP1, ITGA5, ITGAV, ITGB1, and ITGB5 in HCC cells (C), HCC cells silenced for ITGB1 and/or ITGB5 (D), and HCC cells incubated with/without SPP1‐BP (E).Abbreviations: ITGA4, integrin subunit alpha4; ITGA5, integrin subunit alpha5; ITGA8, integrin subunit alpha8; ITGA9, integrin subunit alpha9; ITGAV, integrin subunit alphaV; ITGB1, integrin subunit beta1; ITGB3, integrin subunit beta3; ITGB5, integrin subunit beta; GAPDH, glyceraldehyde 3‐phosphate dehydrogenase; RFU, relative fluorescence units; IgG, immunoglobulin G; SPP1, secreted phosphoprotein 1; CAF, cancer‐associated fibroblast; CM, culture medium; IB, immunoblotting; siCtrl, negative control; SPP1‐BP, SPP1‐blocking peptide. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/36919193), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Osteopontin/OPN by Western Blot
Integrin complexes (integrin alphaV beta5, alpha5 beta1, and alphaV beta1) on HCC cells were identified as binding proteins of SPP1. (A) Western blotting of SPP1 receptor‐binding protein expression in HCC cells. (B) Measurement of CD44 and integrin expression in Hep3B and Huh‐7 cells by fluorescent intensity. (C‐E) Co‐IP of SPP1 after CAF‐CM treatment, followed by Western blotting for SPP1, ITGA5, ITGAV, ITGB1, and ITGB5 in HCC cells (C), HCC cells silenced for ITGB1 and/or ITGB5 (D), and HCC cells incubated with/without SPP1‐BP (E).Abbreviations: ITGA4, integrin subunit alpha4; ITGA5, integrin subunit alpha5; ITGA8, integrin subunit alpha8; ITGA9, integrin subunit alpha9; ITGAV, integrin subunit alphaV; ITGB1, integrin subunit beta1; ITGB3, integrin subunit beta3; ITGB5, integrin subunit beta; GAPDH, glyceraldehyde 3‐phosphate dehydrogenase; RFU, relative fluorescence units; IgG, immunoglobulin G; SPP1, secreted phosphoprotein 1; CAF, cancer‐associated fibroblast; CM, culture medium; IB, immunoblotting; siCtrl, negative control; SPP1‐BP, SPP1‐blocking peptide. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/36919193), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Osteopontin/OPN by Western Blot
Molecular mechanism underlying CAF‐derived SPP1‐induced resistance to sorafenib or lenvatinib. (A‐B) Changes in the expression levels of PKC alpha, BRAF/ERK/STAT3, and PI3K/AKT/mTOR pathway proteins in CAF‐CM‐incubated Huh‐7 cells treated with sorafenib (15 μmol/L; A) or lenvatinib alone (5 μmol/L; B) and in combination with SPP1‐BP. (C‐D) Changes in the phosphorylation of AKT, mTOR, BRAF, and ERK1/2 under ITGB1 and/or ITGB5 silencing in Hep3B and Huh‐7 cells treated with sorafenib (C) or lenvatinib (D) in combination with CAF‐CM. (Mean ± SEM; Two‐way ANOVA test; *P < 0.05; **P < 0.01; ***P < 0.001).Abbreviations: DMSO, dimethyl sulfoxide; CAF, cancer‐associated fibroblast; CM, culture medium; Sor, sorafenib; SPP1‐BP, SPP1‐blocking peptide, PKC alpha, protein kinase C alpha; BRAF,v‐Raf murine sarcoma viral oncogene homolog B; ERK, extracellular signal‐related kinase; STAT3, signal transducer and activator of transcription 3; GAPDH, glyceraldehyde 3‐phosphate dehydrogenase; PI3K, phosphatidylinositol‐3‐kinase; AKT, protein kinase B; mTOR, mammalian target of rapamycin; Len, lenvatinib; siCtrl, negative control; ITGB1, integrin subunit beta1; ITGB5, integrin subunit beta5. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/36919193), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Osteopontin/OPN by Western Blot
Integrin complexes (integrin alphaV beta5, alpha5 beta1, and alphaV beta1) on HCC cells were identified as binding proteins of SPP1. (A) Western blotting of SPP1 receptor‐binding protein expression in HCC cells. (B) Measurement of CD44 and integrin expression in Hep3B and Huh‐7 cells by fluorescent intensity. (C‐E) Co‐IP of SPP1 after CAF‐CM treatment, followed by Western blotting for SPP1, ITGA5, ITGAV, ITGB1, and ITGB5 in HCC cells (C), HCC cells silenced for ITGB1 and/or ITGB5 (D), and HCC cells incubated with/without SPP1‐BP (E).Abbreviations: ITGA4, integrin subunit alpha4; ITGA5, integrin subunit alpha5; ITGA8, integrin subunit alpha8; ITGA9, integrin subunit alpha9; ITGAV, integrin subunit alphaV; ITGB1, integrin subunit beta1; ITGB3, integrin subunit beta3; ITGB5, integrin subunit beta; GAPDH, glyceraldehyde 3‐phosphate dehydrogenase; RFU, relative fluorescence units; IgG, immunoglobulin G; SPP1, secreted phosphoprotein 1; CAF, cancer‐associated fibroblast; CM, culture medium; IB, immunoblotting; siCtrl, negative control; SPP1‐BP, SPP1‐blocking peptide. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/36919193), licensed under a CC-BY license. Not internally tested by R&D Systems.
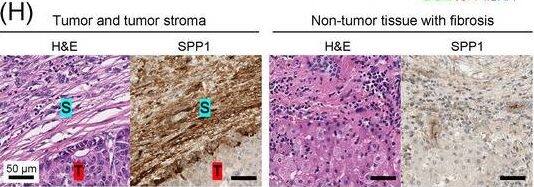
Detection of Osteopontin/OPN by Immunohistochemistry
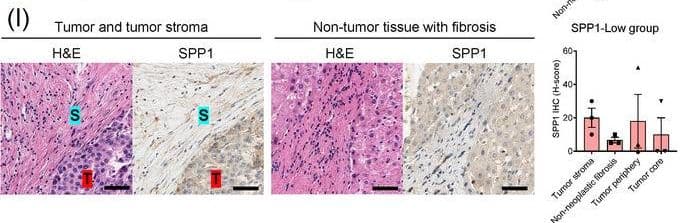
Identification of the CAF‐derived molecules that induce resistance to sorafenib or lenvatinib in patients with HCC. (A) Heat map of 790 genes overexpressed in CAFs. (B) GSEA of the genes overexpressed in CAFs. (C) Process of candidate gene selection. (D) Expression levels of the 9 candidate genes according to sorafenib response in the GSE109211 dataset. (E) Comparison of SPP1 expression between sorafenib responders and non‐responders in the GSE109211 and GSE143233 datasets. (F) Immunofluorescence staining of alpha‐SMA and SPP1 in tumor tissues from patients with HCC. (G) Comparison of SPP1 expression between CAFs and their paired para‐cancer fibroblasts in the WTS data from the 9 pairs of CAF and para‐cancer fibroblasts. (H) Representative H&E and SPP1 IHC images of the tumor (T), tumor stroma (S) (left), and non‐tumor fibrous tissue (middle) in an SPP1‐high patient. Scale bar, 50 μm. Comparison of SPP1 IHC staining intensity according to tumor location in the SPP1‐high group (right). (I) Representative H&E and SPP1 IHC images of tumor (T), tumor stroma (S) (left), and non‐tumor fibrous tissue (middle) in an SPP1‐low patient. Comparison of SPP1 IHC staining intensity according to tumor location in the SPP1‐low group (right). (Mean ± SEM; unpaired Welch's t‐test; *P < 0.05; **P < 0.01; ***P < 0.001).Abbreviations: HCC, hepatocellular carcinoma; NF, normal fibroblast; CAFs, cancer‐associated fibroblasts; ELISA, enzyme linked immunosorbent assay; H&E, hematoxylin and eosin; alpha‐SMA, alpha smooth muscle actin; SPP1, secreted phosphoprotein 1; DAPI, 4′, 6‐diamidino‐2‐phenylindole; PAF, para‐cancer fibroblast; S, tumor stroma; T, tumor; IHC, immunohistochemistry. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/36919193), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Osteopontin/OPN by Western Blot
Integrin complexes (integrin alphaV beta5, alpha5 beta1, and alphaV beta1) on HCC cells were identified as binding proteins of SPP1. (A) Western blotting of SPP1 receptor‐binding protein expression in HCC cells. (B) Measurement of CD44 and integrin expression in Hep3B and Huh‐7 cells by fluorescent intensity. (C‐E) Co‐IP of SPP1 after CAF‐CM treatment, followed by Western blotting for SPP1, ITGA5, ITGAV, ITGB1, and ITGB5 in HCC cells (C), HCC cells silenced for ITGB1 and/or ITGB5 (D), and HCC cells incubated with/without SPP1‐BP (E).Abbreviations: ITGA4, integrin subunit alpha4; ITGA5, integrin subunit alpha5; ITGA8, integrin subunit alpha8; ITGA9, integrin subunit alpha9; ITGAV, integrin subunit alphaV; ITGB1, integrin subunit beta1; ITGB3, integrin subunit beta3; ITGB5, integrin subunit beta; GAPDH, glyceraldehyde 3‐phosphate dehydrogenase; RFU, relative fluorescence units; IgG, immunoglobulin G; SPP1, secreted phosphoprotein 1; CAF, cancer‐associated fibroblast; CM, culture medium; IB, immunoblotting; siCtrl, negative control; SPP1‐BP, SPP1‐blocking peptide. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/36919193), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Osteopontin/OPN by Western Blot
Integrin complexes (integrin alphaV beta5, alpha5 beta1, and alphaV beta1) on HCC cells were identified as binding proteins of SPP1. (A) Western blotting of SPP1 receptor‐binding protein expression in HCC cells. (B) Measurement of CD44 and integrin expression in Hep3B and Huh‐7 cells by fluorescent intensity. (C‐E) Co‐IP of SPP1 after CAF‐CM treatment, followed by Western blotting for SPP1, ITGA5, ITGAV, ITGB1, and ITGB5 in HCC cells (C), HCC cells silenced for ITGB1 and/or ITGB5 (D), and HCC cells incubated with/without SPP1‐BP (E).Abbreviations: ITGA4, integrin subunit alpha4; ITGA5, integrin subunit alpha5; ITGA8, integrin subunit alpha8; ITGA9, integrin subunit alpha9; ITGAV, integrin subunit alphaV; ITGB1, integrin subunit beta1; ITGB3, integrin subunit beta3; ITGB5, integrin subunit beta; GAPDH, glyceraldehyde 3‐phosphate dehydrogenase; RFU, relative fluorescence units; IgG, immunoglobulin G; SPP1, secreted phosphoprotein 1; CAF, cancer‐associated fibroblast; CM, culture medium; IB, immunoblotting; siCtrl, negative control; SPP1‐BP, SPP1‐blocking peptide. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/36919193), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Osteopontin/OPN by Immunohistochemistry
Identification of the CAF‐derived molecules that induce resistance to sorafenib or lenvatinib in patients with HCC. (A) Heat map of 790 genes overexpressed in CAFs. (B) GSEA of the genes overexpressed in CAFs. (C) Process of candidate gene selection. (D) Expression levels of the 9 candidate genes according to sorafenib response in the GSE109211 dataset. (E) Comparison of SPP1 expression between sorafenib responders and non‐responders in the GSE109211 and GSE143233 datasets. (F) Immunofluorescence staining of alpha‐SMA and SPP1 in tumor tissues from patients with HCC. (G) Comparison of SPP1 expression between CAFs and their paired para‐cancer fibroblasts in the WTS data from the 9 pairs of CAF and para‐cancer fibroblasts. (H) Representative H&E and SPP1 IHC images of the tumor (T), tumor stroma (S) (left), and non‐tumor fibrous tissue (middle) in an SPP1‐high patient. Scale bar, 50 μm. Comparison of SPP1 IHC staining intensity according to tumor location in the SPP1‐high group (right). (I) Representative H&E and SPP1 IHC images of tumor (T), tumor stroma (S) (left), and non‐tumor fibrous tissue (middle) in an SPP1‐low patient. Comparison of SPP1 IHC staining intensity according to tumor location in the SPP1‐low group (right). (Mean ± SEM; unpaired Welch's t‐test; *P < 0.05; **P < 0.01; ***P < 0.001).Abbreviations: HCC, hepatocellular carcinoma; NF, normal fibroblast; CAFs, cancer‐associated fibroblasts; ELISA, enzyme linked immunosorbent assay; H&E, hematoxylin and eosin; alpha‐SMA, alpha smooth muscle actin; SPP1, secreted phosphoprotein 1; DAPI, 4′, 6‐diamidino‐2‐phenylindole; PAF, para‐cancer fibroblast; S, tumor stroma; T, tumor; IHC, immunohistochemistry. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/36919193), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Osteopontin/OPN by Immunohistochemistry
Identification of the CAF‐derived molecules that induce resistance to sorafenib or lenvatinib in patients with HCC. (A) Heat map of 790 genes overexpressed in CAFs. (B) GSEA of the genes overexpressed in CAFs. (C) Process of candidate gene selection. (D) Expression levels of the 9 candidate genes according to sorafenib response in the GSE109211 dataset. (E) Comparison of SPP1 expression between sorafenib responders and non‐responders in the GSE109211 and GSE143233 datasets. (F) Immunofluorescence staining of alpha‐SMA and SPP1 in tumor tissues from patients with HCC. (G) Comparison of SPP1 expression between CAFs and their paired para‐cancer fibroblasts in the WTS data from the 9 pairs of CAF and para‐cancer fibroblasts. (H) Representative H&E and SPP1 IHC images of the tumor (T), tumor stroma (S) (left), and non‐tumor fibrous tissue (middle) in an SPP1‐high patient. Scale bar, 50 μm. Comparison of SPP1 IHC staining intensity according to tumor location in the SPP1‐high group (right). (I) Representative H&E and SPP1 IHC images of tumor (T), tumor stroma (S) (left), and non‐tumor fibrous tissue (middle) in an SPP1‐low patient. Comparison of SPP1 IHC staining intensity according to tumor location in the SPP1‐low group (right). (Mean ± SEM; unpaired Welch's t‐test; *P < 0.05; **P < 0.01; ***P < 0.001).Abbreviations: HCC, hepatocellular carcinoma; NF, normal fibroblast; CAFs, cancer‐associated fibroblasts; ELISA, enzyme linked immunosorbent assay; H&E, hematoxylin and eosin; alpha‐SMA, alpha smooth muscle actin; SPP1, secreted phosphoprotein 1; DAPI, 4′, 6‐diamidino‐2‐phenylindole; PAF, para‐cancer fibroblast; S, tumor stroma; T, tumor; IHC, immunohistochemistry. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/36919193), licensed under a CC-BY license. Not internally tested by R&D Systems.
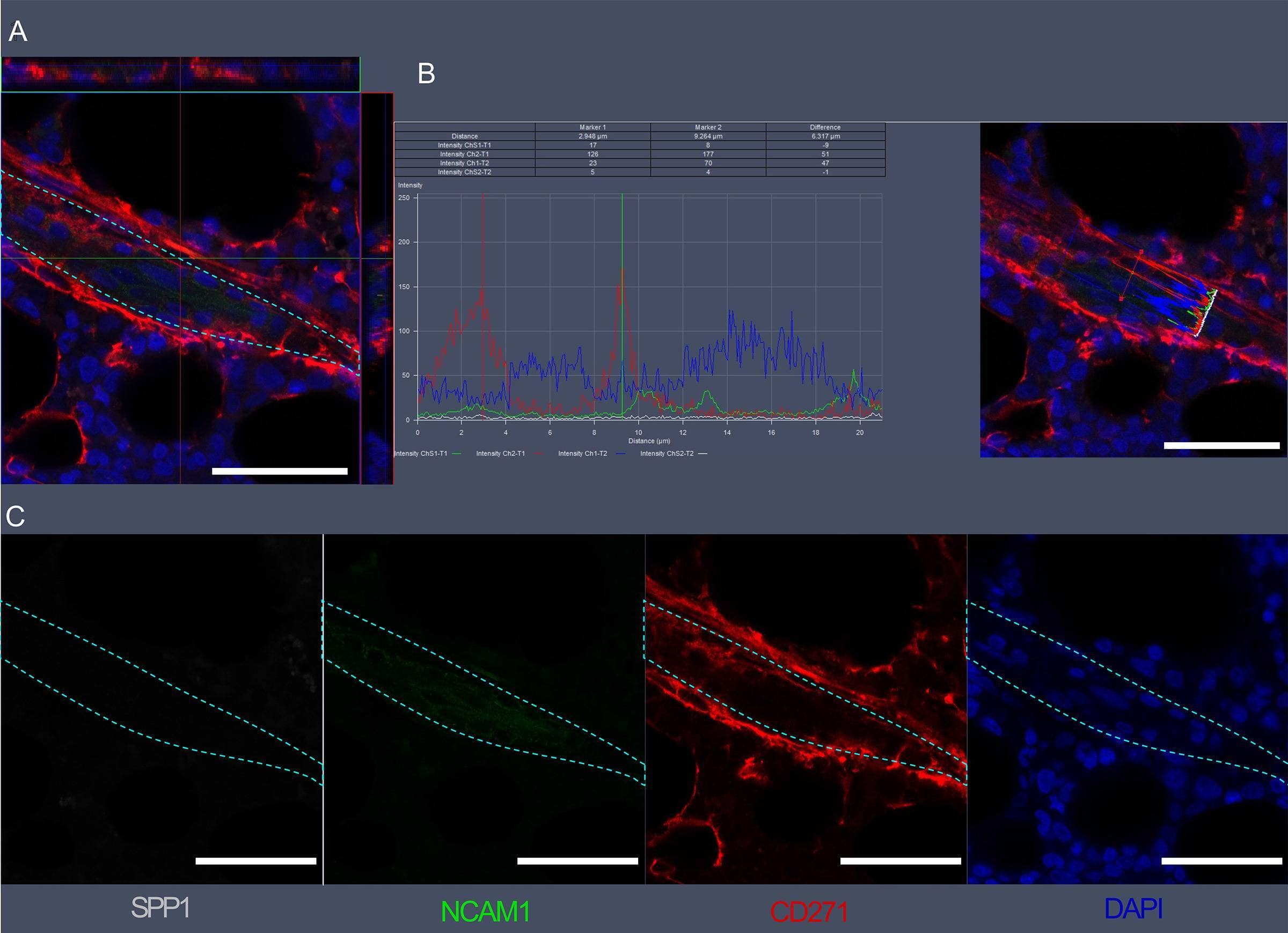
Detection of Osteopontin/OPN by Immunohistochemistry
Visualization of SPP1, NCAM1, and CD271 expression in bone marrow vascular regions by confocal microscopy.(A) Confocal scan of vascular region in BM biopsies with 3D orthographic cross-section view, co-stained with rabbit anti-NCAM1, mouse anti-CD271, goat anti-SPP1, and DAPI. (B) Intensity profile for all channels in A across a cell of interest in a representative z plane. (C) Single channel data for the florescent markers in A. Scale bars represent 50 μm. Cyan dashed lines indicate vessel surface. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/36876630), licensed under a CC-BY license. Not internally tested by R&D Systems.
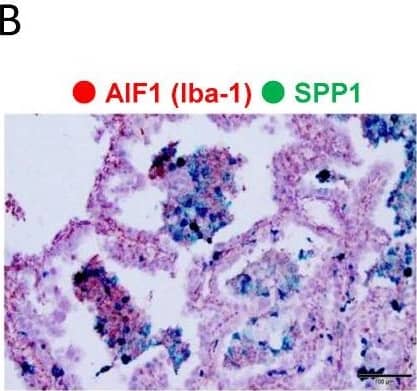
Detection of Osteopontin/OPN by Immunohistochemistry
Evaluation of SPP1 expression on cancer cells and macrophages. (A,B). Representative dual ISH (green: SPP1; red: Iba-1) in adenocarcinoma. (C). Representative single-IHC of SPP1 in adenocarcinoma and SCC. (D). Double-IHC of SPP1 and Iba-1 to examine the SPP1 expression level in cancer cells (Iba-1-negative). Double-IHC of SPP1 and PU.1 to examine the SPP1 expression level on TAMs (PU.1-positive in the nucleus). (E). SPP1’s expression level was scored according to the proportion of stained cells as follows: less than 1% staining, score 0; 1% to 49% staining, score 1; more than 50% staining, score 2. The proportions of samples with each score among cancer cells and TAMs in adenocarcinoma and SCC are shown in the pie charts. Image collected and cropped by CiteAb from the following open publication (https://www.mdpi.com/2072-6694/14/18/4374), licensed under a CC-BY license. Not internally tested by R&D Systems.
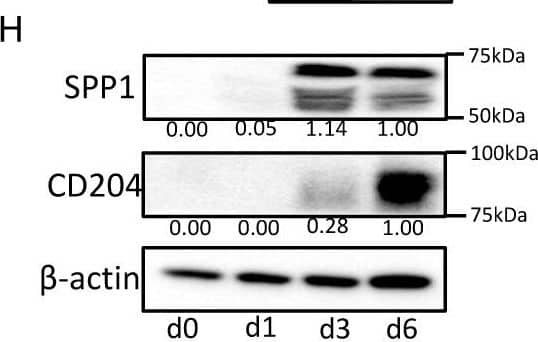
Detection of Osteopontin/OPN by Western Blot
SPP1 expression in macrophages. (A) SPP1 expression in macrophages, THP-1 cells, U937 cells, and 10 lung cancer cell lines was examined by qRT-PCR. (B) Macrophages (blue) and THP-1 cells (green) were stimulated with the CM of cancer cell lines; then, the SPP1 mRNA expression level was evaluated. Control (CT) refers to macrophages or THP-1 without CM of cancer cell lines added. (C) Monocytes were cultured with M-CSF and GM-CSF as described in the Materials and Methods section, and SPP1 mRNA and CD204 mRNA levels were evaluated at different stages of monocyte/macrophage culture (days 0, 3, and 5). (D) Monocytes were cultured with M-CSF or GM-CSF for 5 days, and SPP1 and CD204 expression levels were examined. (E) SPP1′s concentration (ng/mL) in the CM of macrophages cultured with M-CSF or GM-CSF was examined by ELISA at days 1 to 3 (n = 3). (F) Immunocytostaining of SPP1 in the macrophages cultured with M-CSF or GM-CSF at days 0 and 5. (G) SPP1 and CD204 expression levels in THP-1 cells differentiated for 0, 1, 3, or 6 days were examined by qRT-PCR (G) and Western blot analysis (H). *: statistically significant, p value < 0.05. Image collected and cropped by CiteAb from the following open publication (https://www.mdpi.com/2072-6694/14/18/4374), licensed under a CC-BY license. Not internally tested by R&D Systems.