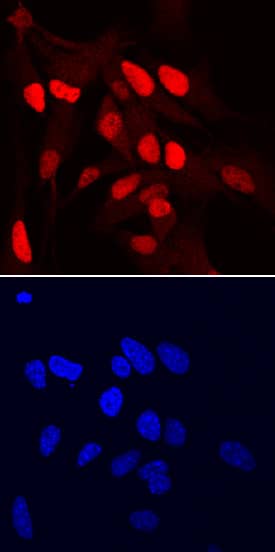
SOX1 in Rat Cortical Stem Cells.
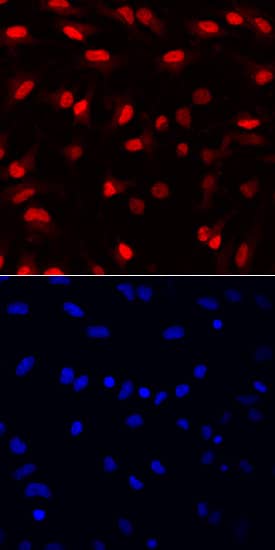
SOX1 was detected in immersion fixed rat cortical stem cells (
NSC001) using Goat Anti-Human/Mouse/Rat SOX1 Antigen Affinity-purified Polyclonal Antibody (Catalog # AF3369) at 10 µg/mL for 3 hours at room temperature. Cells were stained using the NorthernLights™ 557-conjugated Anti-Goat IgG Secondary Antibody (red, upper panel;
NL001) and counterstained with DAPI (blue, lower panel). Specific staining was localized to nuclei. View our protocol for Fluorescent ICC Staining of Cells on Coverslips.
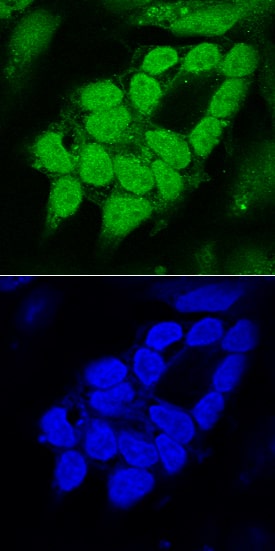
SOX1 in Mouse Cortical Stem Cells.
SOX1 was detected in immersion fixed mouse cortical stem cells (Catalog #
NSC002) using Goat Anti-Human/Mouse/Rat SOX1 Antigen Affinity-purified Polyclonal Antibody (Catalog # AF3369) at 10 µg/mL for 3 hours at room temperature. Cells were stained using the NorthernLights™ 557-conjugated Anti-Goat IgG Secondary Antibody (red, upper panel;
NL001) and counterstained with DAPI (blue, lower panel). Specific staining was localized to nuclei. View our protocol for Fluorescent ICC Staining of Cells on Coverslips.
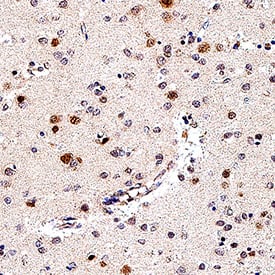
SOX1 in Human Brain (Cortex).
SOX1 was detected in immersion fixed paraffin-embedded sections of human brain (cortex) using Goat Anti-Human/Mouse/Rat SOX1 Antigen Affinity-purified Polyclonal Antibody (Catalog # AF3369) at 3 µg/mL for 1 hour at room temperature followed by incubation with the Anti-Mouse IgG VisUCyte™ HRP Polymer Antibody (
VC001). Before incubation with the primary antibody, tissue was subjected to heat-induced epitope retrieval using Antigen Retrieval Reagent-Basic (
CTS013). Tissue was stained using DAB (brown) and counterstained with hematoxylin (blue). Specific staining was localized to nuclei in neuron. Staining was performed using our protocol for IHC Staining with VisUCyte HRP Polymer Detection Reagents.
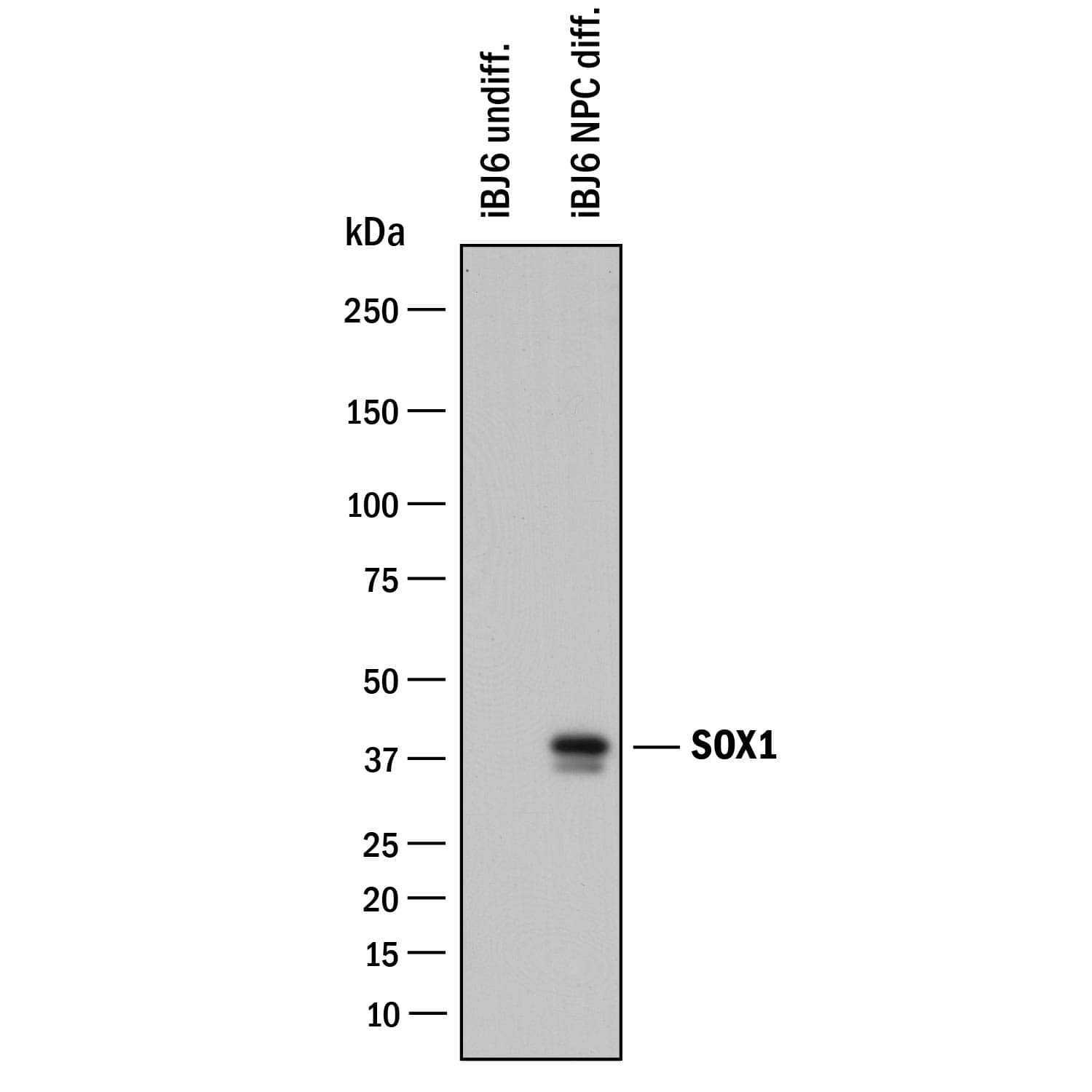
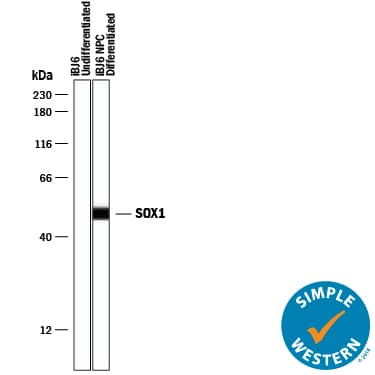
Detection of Human SOX1 by Simple WesternTM.
Simple Western lane view shows lysates of undifferentiated iBJ6 human iPS cells and iBJ6 human iPS cells differentiated into neuroprogenitor cells, loaded at 0.2 mg/mL. A specific band was detected for SOX1 at approximately 50 kDa (as indicated) using 10 µg/mL of Goat Anti-Human/Mouse/Rat SOX1 Antigen Affinity-purified Polyclonal Antibody (Catalog # AF3369) followed by 1:50 dilution of HRP-conjugated Anti-Goat IgG Secondary Antibody (
HAF109). This experiment was conducted under reducing conditions and using the 12-230 kDa separation system.
Detection of Human SOX1 by Immunocytochemistry/Immunofluorescence
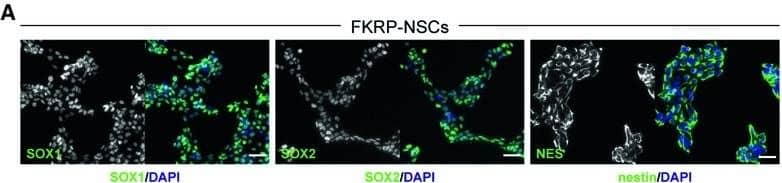
Characterization of NSCs and cortical neurons derived from FKRP‐ and CRISPR/Cas9 corrected‐iPSCsA, BRepresentative images of NSCs derived from FKRP‐ and corrected‐iPSC lines expressing SOX1, SOX2, and nestin.C, DQuantification of percentage of SOX1+ (C) and SOX2+ (D) cells in culture. The efficiency of neural induction is more than 99% in FKRP‐ and corrected‐iPSC (5D17, 5D23, and 3B17) lines. Data are mean ± s.d. n = 4 technical replicates.E, FFKRP‐ and corrected‐NSC lines can be further differentiated to cortical neural progenitor cells, expressing PAX6, OTX2, and vimentin.G–IQuantification of percentage of PAX6+ (G) and OTX2+ (H) cells in culture. About 91‐98% of cells derived from FKRP, 5D17, 5D23, and 3B17 NSC lines express PAX6 (G). About 93‐96% of cells derived from FKRP, 5D17, 5D23, and 3B17 NSC lines express OTX2 (H). Of the OTX2+ population, about 60‐67% cells are also Ki67+ cycling progenitors (I). Data are mean ± s.d. n = 4 technical replicates.J, KGlutamatergic projection neurons derived from FKRP and corrected (5D17, 5D23, and 3B17) progenitor cells. The vast majority of neurons contain vGlut1+ punctae in their neurites (labeled by Tuj1). Right panels are enlarged images from the insets of left panels.Data information: Scale bars, 50 μm. Image collected and cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/31566294), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Human SOX1 by Immunocytochemistry/Immunofluorescence
Characterization of NSCs and cortical neurons derived from FKRP‐ and CRISPR/Cas9 corrected‐iPSCsA, BRepresentative images of NSCs derived from FKRP‐ and corrected‐iPSC lines expressing SOX1, SOX2, and nestin.C, DQuantification of percentage of SOX1+ (C) and SOX2+ (D) cells in culture. The efficiency of neural induction is more than 99% in FKRP‐ and corrected‐iPSC (5D17, 5D23, and 3B17) lines. Data are mean ± s.d. n = 4 technical replicates.E, FFKRP‐ and corrected‐NSC lines can be further differentiated to cortical neural progenitor cells, expressing PAX6, OTX2, and vimentin.G–IQuantification of percentage of PAX6+ (G) and OTX2+ (H) cells in culture. About 91‐98% of cells derived from FKRP, 5D17, 5D23, and 3B17 NSC lines express PAX6 (G). About 93‐96% of cells derived from FKRP, 5D17, 5D23, and 3B17 NSC lines express OTX2 (H). Of the OTX2+ population, about 60‐67% cells are also Ki67+ cycling progenitors (I). Data are mean ± s.d. n = 4 technical replicates.J, KGlutamatergic projection neurons derived from FKRP and corrected (5D17, 5D23, and 3B17) progenitor cells. The vast majority of neurons contain vGlut1+ punctae in their neurites (labeled by Tuj1). Right panels are enlarged images from the insets of left panels.Data information: Scale bars, 50 μm. Image collected and cropped by CiteAb from the following publication (https://pubmed.ncbi.nlm.nih.gov/31566294), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Human SOX1 by Immunocytochemistry/Immunofluorescence
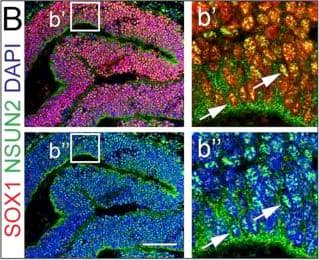
Expression of NSUN2 in the Human Developing Brain and NES Cells(A) DAPI-stained human embryo (6 weeks of gestation) marked for prosencephalon, mesencephalon, and rhombencephalon. Region in square is magnified in (B). Scale bar, 1 mm.(B) Prosencephalon labeled for NSUN2 and SOX1. Region in squares are magnified in (b′) and (b″). Arrows indicate NSUN2-positive cells. Scale bar, 100 μm.(C–F) Bright-field image (C) and immunofluorescence (D–F) of AF22 (upper panels) and Sai1 (lower panels) cells labeled for Nestin (D), SOX2 (E), and betaIII-tubulin (F). Scale bar, 50 μm.(G and H) NES cells co-labeled for NSUN2 and Nestin (NES) (G) or SOX1 (H).(I) Differentiation protocol.(J–L) Differentiated AF22 and Sai1 cells (day 15) labeled for Nestin (NES; J), SOX2 (K), and betaIII-tubulin (L). Scale bars: 50 μm.(M) Western blot for NSUN2, betaIII-tubulin (TUBB3), GFAP, SOX2, and Nestin during differentiation (days). alpha-Tubulin served as loading control.Nuclei are counterstained with DAPI (A, B, D–F, J–L). Image collected and cropped by CiteAb from the following publication (https://linkinghub.elsevier.com/retrieve/pii/S2213671116302764), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Human SOX1 by Immunocytochemistry/Immunofluorescence
Expression of NSUN2 in the Human Developing Brain and NES Cells(A) DAPI-stained human embryo (6 weeks of gestation) marked for prosencephalon, mesencephalon, and rhombencephalon. Region in square is magnified in (B). Scale bar, 1 mm.(B) Prosencephalon labeled for NSUN2 and SOX1. Region in squares are magnified in (b′) and (b″). Arrows indicate NSUN2-positive cells. Scale bar, 100 μm.(C–F) Bright-field image (C) and immunofluorescence (D–F) of AF22 (upper panels) and Sai1 (lower panels) cells labeled for Nestin (D), SOX2 (E), and betaIII-tubulin (F). Scale bar, 50 μm.(G and H) NES cells co-labeled for NSUN2 and Nestin (NES) (G) or SOX1 (H).(I) Differentiation protocol.(J–L) Differentiated AF22 and Sai1 cells (day 15) labeled for Nestin (NES; J), SOX2 (K), and betaIII-tubulin (L). Scale bars: 50 μm.(M) Western blot for NSUN2, betaIII-tubulin (TUBB3), GFAP, SOX2, and Nestin during differentiation (days). alpha-Tubulin served as loading control.Nuclei are counterstained with DAPI (A, B, D–F, J–L). Image collected and cropped by CiteAb from the following publication (https://linkinghub.elsevier.com/retrieve/pii/S2213671116302764), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Human SOX1 by Immunocytochemistry/Immunofluorescence
GPI anchored proteins are required for neural differentiation.(A). Representative example of images of hiPSC-derived EBs and EB-derived rosettes during neural differentiation. Neural induction and rosette formation upon neural induction was assessed in three cell lines using a serum-free EB generation method. On day 2 (left) of hiPSC-derived EBs from PIGAwt, PIGAc.1234C>T, and PIGAnull after forced aggregation (20X magnification, scale bar is 50μm). On day 4 (middle), single homogeneous hiPSC-EBs collected were pooled in a 10 cm plate (4X magnification, scale bar is 100μm). On day 11 (right), neuroepithelial cells appeared and neural tube-like rosettes formed (EB-derived rosettes) and scale bar is 50μm. (B). Neural induction rates from EB-derived rosettes. The percentage of EB derived rosettes was 88.8% ± 4.6, 75.5% ± 9.8 and 68.4% ± 6.9 for PIGAwt, PIGAc.1234C>T, and PIGAnull, respectively. PIGAwt versus PIGAc.1234C>T (p>0.05, NS) and PIGAwt versus PIGAnull (*p<0.05, one way ANOVA and Multiple comparisons). Neural induction from PIGAnull hiPSCs was less than 70%. All values were mean ±SD. (C). Representative confocal images showing expression of neuron stem cell marker SOX1 (in red) combined proliferation by EdU labeling in hNPCs derived from isolated neural rosettes. Nuclei were visualized with DAPI (blue) and scale bar 100μm. (D). Representative confocal images showing expression of neuron progenitor marker PAX6 (in red) and combined proliferation by EdU (in green) in hNPCs derived from isolated neural rosettes. Nuclei were visualized with DAPI (blue) and scale bar 200μm. hNPCs from PIGAnull cell lines showed reduced expression of SOX1 and PAX6. (E). Proliferation rate in hNPCs was assessed and plotted in all three cell lines. EdU positive cells were counted and normalized by total number of nuclei staining with DAPI (blue). Proliferation was significantly decreased in PIGAnull and PIGAc.1234C>T compared to PIGAwt. (F). Graphs depict the percentage of positive cells for SOX-1 (left) and Pax6 (right) in hNPCs derived from PIGAwt, PIGAc.1234C>T, and PIGAnull hiPSC lines. The hNPCs derived from the PIGAnull hiPSCs showed significantly decreased expression of SOX1 and PAX6. Similar levels of SOX1 and PAX6 were expressed in hNPCs from PIGAwt and PIGAc.1234C>T. All values represent mean ± SD. Image collected and cropped by CiteAb from the following publication (https://dx.plos.org/10.1371/journal.pone.0174074), licensed under a CC-BY license. Not internally tested by R&D Systems.
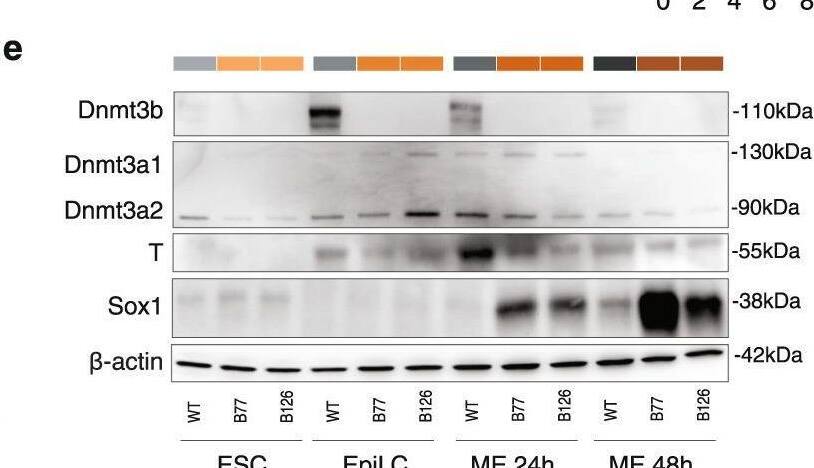
Detection of SOX1 by Western Blot
Loss of DNMT3B impairs meso-endoderm lineage commitment. A Schematic representation of the two-step differentiation model from ESCs to EpiLCs first with Fgf beta & Activin A, & then to meso-endoderm (ME) progenitors with iGsk3. The time points of ’ collection are reported in the colour-code used throughout the figures (i.e., shades of grey for WT, shades of orange for 3BKO). b Hierarchical clustering of RNA-seq data from the in vitro differentiation & in vivo embryonic tissues derived from pre- & post-implantation mouse embryos35. Pearson’s correlation distance & Ward’s method employed to perform the analysis. c On the left, an RNA-seq heatmap showing the results of gene expression profiles clustering with K-means for WT & 3BKO (two independent clones) during the complete differentiation time course (ESC-EpiLC-ME). DEGs arising during the differentiation time course in any group identified by ANOVA-like test with edgeR54. Rows are genes, columns are samples & the scaled expression level (Z-score RPKM) is plotted. On the right, heatmap showing selected GO terms for enriched biological processes in each cluster. Terms related to meso-endoderm are highlighted in red. d Gene expression time-course for stage-specific pluripotency (naive, primed) & germ layers (mesoderm, endoderm, ectoderm) marker genes. Dots represent normalized RPKM values, averaged by replicates/conditions (n = 2 biological replicates per genotype or clone at each time point). Error bars represent standard errors. e, WB analysis of the de novo DNMTs (Dnmt3a1, Dnmt3a2, Dnmt3b), T (mesodermal marker) & Sox1 (neuro-ectodermal marker) expression during the differentiation time course. beta-actin serves as loading control. Representative of two independent experiments. Uncropped gels are provided in Supplementary Fig. 11. Image collected & cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/36690616), licensed under a CC-BY license. Not internally tested by R&D Systems.
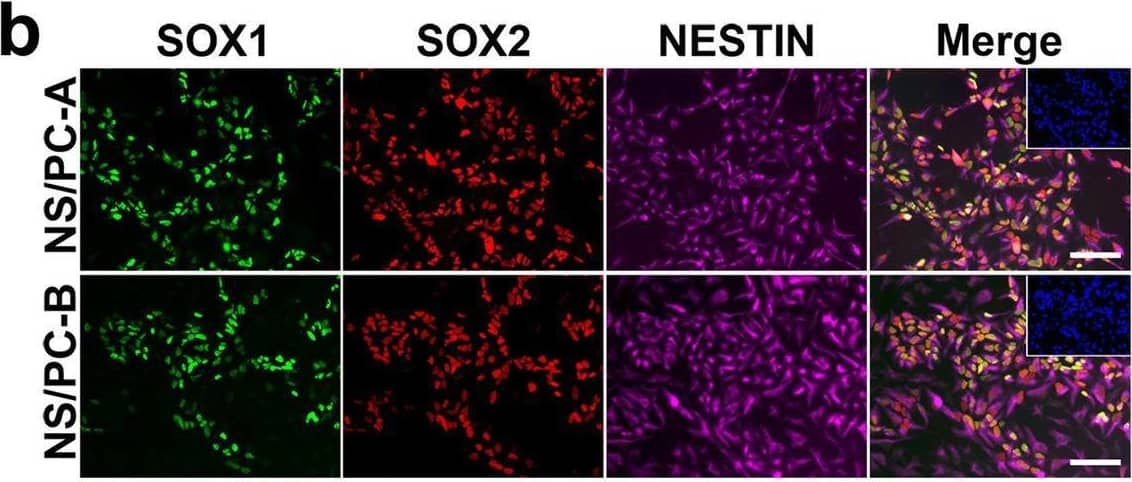
Detection of Human SOX1 by Immunocytochemistry/ Immunofluorescence
Characterization of NS/PCs derived from hiPSCs. b, Representative images (b) of immunocytochemical analysis of hiPSC-NS/PCs (NS/PC-A & NS/PC-B) using antibodies against SOX1, SOX2, & NESTIN. Inset: Hoechst nuclear staining of the same field. Scale bar, 50 μm. Image collected & cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/37286713), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of SOX1 by Western Blot
Loss of DNMT3B impairs meso-endoderm lineage commitment. A Schematic representation of the two-step differentiation model from ESCs to EpiLCs first with Fgf beta & Activin A, & then to meso-endoderm (ME) progenitors with iGsk3. The time points of ’ collection are reported in the colour-code used throughout the figures (i.e., shades of grey for WT, shades of orange for 3BKO). b Hierarchical clustering of RNA-seq data from the in vitro differentiation & in vivo embryonic tissues derived from pre- & post-implantation mouse embryos35. Pearson’s correlation distance & Ward’s method employed to perform the analysis. c On the left, an RNA-seq heatmap showing the results of gene expression profiles clustering with K-means for WT & 3BKO (two independent clones) during the complete differentiation time course (ESC-EpiLC-ME). DEGs arising during the differentiation time course in any group identified by ANOVA-like test with edgeR54. Rows are genes, columns are samples & the scaled expression level (Z-score RPKM) is plotted. On the right, heatmap showing selected GO terms for enriched biological processes in each cluster. Terms related to meso-endoderm are highlighted in red. d Gene expression time-course for stage-specific pluripotency (naive, primed) & germ layers (mesoderm, endoderm, ectoderm) marker genes. Dots represent normalized RPKM values, averaged by replicates/conditions (n = 2 biological replicates per genotype or clone at each time point). Error bars represent standard errors. e, WB analysis of the de novo DNMTs (Dnmt3a1, Dnmt3a2, Dnmt3b), T (mesodermal marker) & Sox1 (neuro-ectodermal marker) expression during the differentiation time course. beta-actin serves as loading control. Representative of two independent experiments. Uncropped gels are provided in Supplementary Fig. 11. Image collected & cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/36690616), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Human SOX1 by Immunocytochemistry/ Immunofluorescence
Characterization of NS/PCs derived from hiPSCs. b, Representative images (b) of immunocytochemical analysis of hiPSC-NS/PCs (NS/PC-A & NS/PC-B) using antibodies against SOX1, SOX2, & NESTIN. Inset: Hoechst nuclear staining of the same field. Scale bar, 50 μm. Image collected & cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/37286713), licensed under a CC-BY license. Not internally tested by R&D Systems.