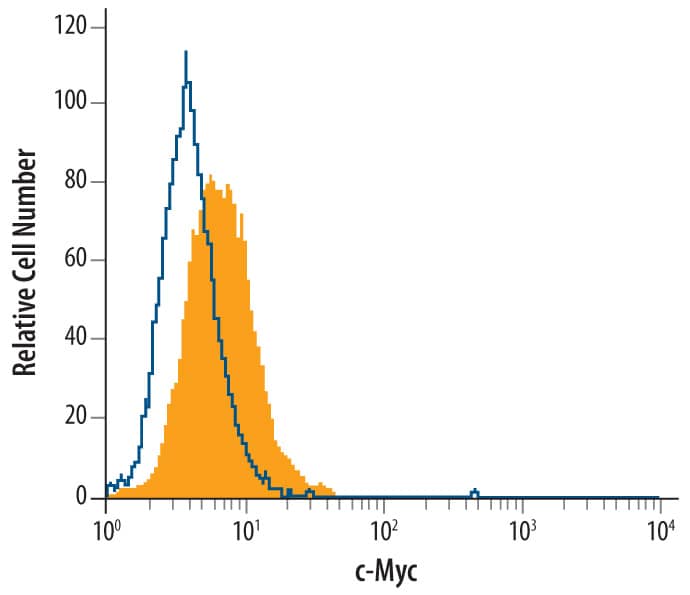
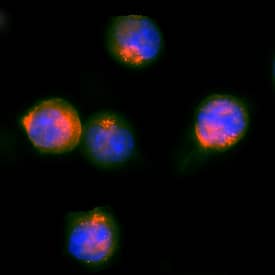
c-Myc in HEK293 Human Cell Line Transfected with c-Myc-tagged Serotonin Receptor.
c-Myc was detected in immersion fixed HEK293 human embryonic kidney cell line transfected with c-Myc-tagged Serotonin Receptor using Mouse Anti-Human c-Myc Monoclonal Antibody (Catalog # MAB3696) at 25 µg/mL for 3 hours at room temperature. Cells were stained using the NorthernLights™ 557-conjugated Anti-Mouse IgG Secondary Antibody (red; Catalog #
NL007) and counterstained with DAPI (blue). Specific staining was localized to nuclei. View our protocol for Fluorescent ICC Staining of Cells on Coverslips.
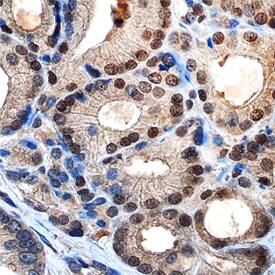
c‑Myc in Human Prostate.
c-Myc was detected in immersion fixed paraffin-embedded sections of human prostate using Mouse Anti-Human c-Myc Monoclonal Antibody (Catalog # MAB3696) at 3 µg/mL overnight at 4 °C. Tissue was stained using the Anti-Mouse HRP-DAB Cell & Tissue Staining Kit (brown; Catalog #
CTS002) and counterstained with hematoxylin (blue). Specific staining was localized to nuclei of epithelial cells. View our protocol for Chromogenic IHC Staining of Paraffin-embedded Tissue Sections.
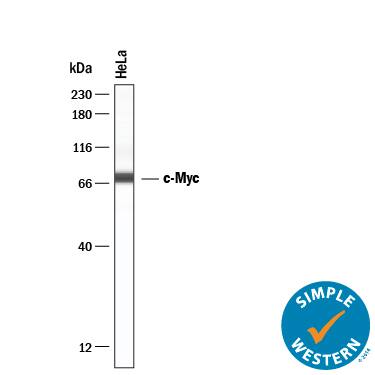
Detection of Human c‑Myc by Simple WesternTM.
Simple Western lane view shows lysates of HeLa human cervical epithelial carcinoma cell line, loaded at 0.2 mg/mL. A specific band was detected for c‑Myc at approximately 74 kDa (as indicated) using 20 µg/mL of Mouse Anti-Human c‑Myc Monoclonal Antibody (Catalog # MAB3696) . This experiment was conducted under reducing conditions and using the 12-230 kDa separation system.
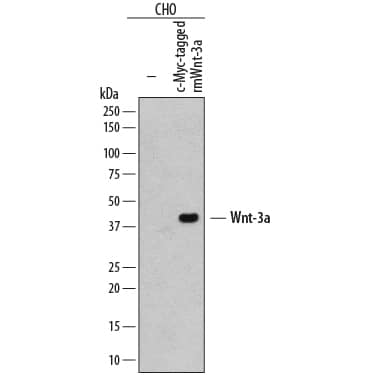
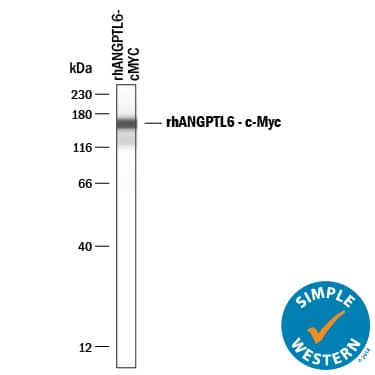
Detection of Human c‑Myc by Simple WesternTM.
Simple Western lane view shows c-Myc-tagged recombinant human ANGPTL6, loaded at 0.2 mg/mL. A specific band was detected for c‑Myc at approximately 161 kDa (as indicated) using 20 µg/mL of Mouse Anti-Human c‑Myc Monoclonal Antibody (Catalog # MAB3696) . This experiment was conducted under reducing conditions and using the 12-230 kDa separation system.
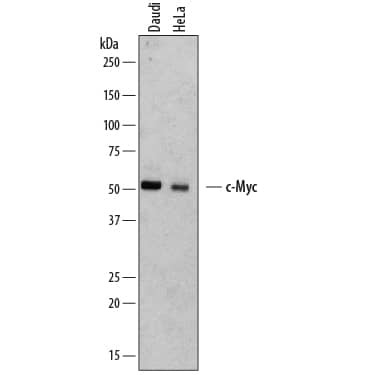
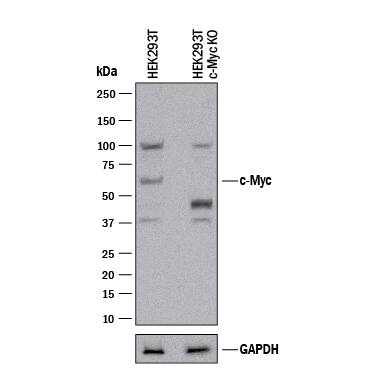
Western Blot Shows Human c‑Myc Specificity by Using Knockout Cell Line.
Western blot shows lysates of HEK293T human embryonic kidney parental cell line and c-Myc knockout HEK293T cell line (KO). PVDF membrane was probed with 2 µg/mL of Mouse Anti-Human c-Myc Monoclonal Antibody (Catalog # MAB3696) followed by HRP-conjugated Anti-Mouse IgG Secondary Antibody (Catalog #
HAF018). A specific band was detected for c-Myc at approximately 52 kDa (as indicated) in the parental HEK293T cell line, but is not detectable in knockout HEK293T cell line. GAPDH (Catalog #
MAB5718) is shown as a loading control. This experiment was conducted under reducing conditions and using Immunoblot Buffer Group 1.
Detection of c-Myc by Western Blot
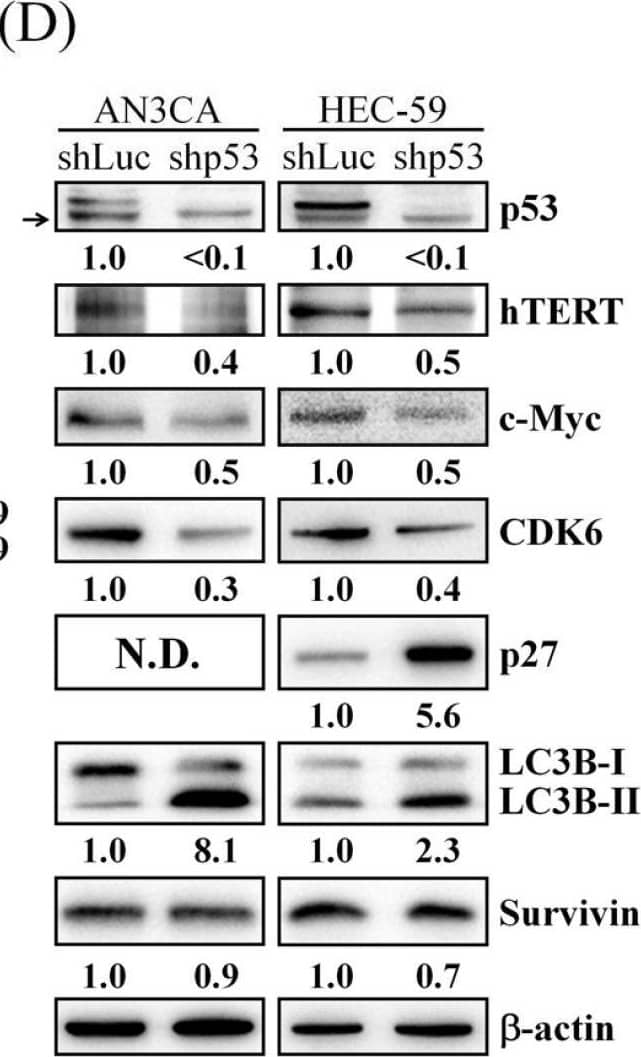
Effect of mtp53 knockdown on cell proliferation and Akt/mTOR pathway. (A) AN3CA shLuc and shp53 cells (2 × 105 cells/35 mm dish) were seeded on a 35 mm dish. After 24 (Day 0), 48 (Day 1), 72 (Day 2), and 96 h (Day 3) incubation, the cells were harvested by trypsin and counted under an inverted microscope. (B) HEC-59 shLuc and shp53 cells (2.5 × 105 cells/35 mm dish) were seeded on a 35 mm dish. After 24 (Day 0), 48 (Day 1), 72 (Day 2), and 96 h (Day 3) incubation, the cells were harvested by trypsin and counted under an inverted microscope. (C) Protein expressions of p53, p-Akt, Akt, p- p-p85S6K/p70S6K, and p85S6K/p70S6K were determined by Western blot. ImageJ software was used to quantify the band intensities of p53, p-Akt, Akt, p-p85S6K/p70S6K, and p85S6K/p70S6K. Data shown are the relative expression standardized by the beta-actin protein level. The ratio of shLuc cells was set at 1. (D) Protein expressions of p53, hTERT, c-Myc, CDK6, p27, LC3B, and survivin were determined by Western blot. The arrow indicated the nonspecific band. ImageJ software was used to quantify the band intensities of indicated protein. Data shown are the relative expression standardized by the beta-actin protein level. The ratio of shLuc cells was set at 1. The symbol ∗ indicates p < 0.05. N.D., not detected. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/34831139), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of c-Myc by Western Blot
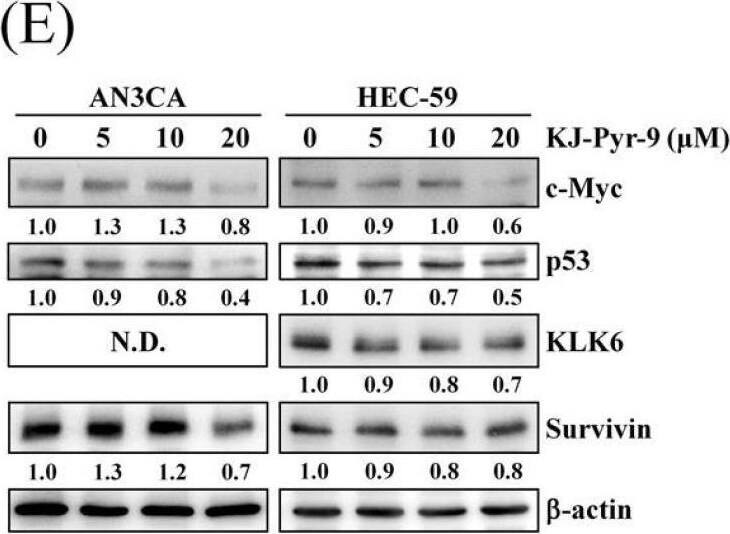
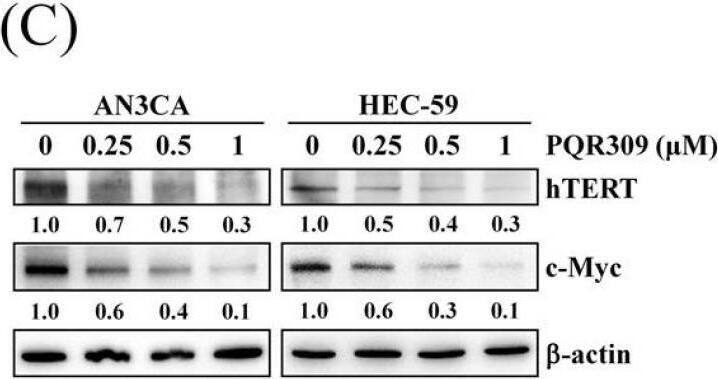
Effect of PQR309 on the expression of c-Myc and its possible target genes. (A) After treating with PQR309 for 48 h in HEC-59 cells, Proteome Profiler Human XL Oncology Array was performed according to the manufacturer protocol. The dot intensities were quantified by ImageJ software. The ratio of cells without treatment was set at 1. (B) After treating with PQR309 in AN3CA and HEC-59 cells for 48 h, protein expressions of endoglin, KLK6, p53, and survivin were determined by Western blot. ImageJ software was used to quantify the band intensities of endoglin, KLK6, p53, and survivin. Data shown are the relative expression standardized by the beta-actin protein level. The ratio of cells without treatment was set at 1. (C) Western blot assay was performed to detect the expressions of hTERT and c-Myc in AN3CA and HEC-59 cells after PQR309 treatment for 48 h. ImageJ software was used to quantify the band intensities of hTERT and c-Myc. Data shown are the relative expression standardized by the beta-actin protein level. The ratio of cells without treatment was set at 1. (D) AN3CA (5 × 103 cells/well of 96-well plate) and HEC-59 (6 × 103 cells/well of 96-well plate) were treated with KJ-Pyr-9 (0, 2.5, 5, 10 and 20 μM), an inhibitor of c-Myc, for 72 h. Cell viability was analyzed by MTT assay. (E) Protein expressions of c-Myc, p53, KLK6, and survivin were determined by Western blot. ImageJ software was used to quantify the band intensities of c-Myc, p53, KLK6, and survivin. Data shown are the relative expression standardized by the beta-actin protein level. The ratio of cells without treatment was set at 1. N.D., not detected. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/34831139), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of c-Myc by Western Blot
Effect of PQR309 on the expression of c-Myc and its possible target genes. (A) After treating with PQR309 for 48 h in HEC-59 cells, Proteome Profiler Human XL Oncology Array was performed according to the manufacturer protocol. The dot intensities were quantified by ImageJ software. The ratio of cells without treatment was set at 1. (B) After treating with PQR309 in AN3CA and HEC-59 cells for 48 h, protein expressions of endoglin, KLK6, p53, and survivin were determined by Western blot. ImageJ software was used to quantify the band intensities of endoglin, KLK6, p53, and survivin. Data shown are the relative expression standardized by the beta-actin protein level. The ratio of cells without treatment was set at 1. (C) Western blot assay was performed to detect the expressions of hTERT and c-Myc in AN3CA and HEC-59 cells after PQR309 treatment for 48 h. ImageJ software was used to quantify the band intensities of hTERT and c-Myc. Data shown are the relative expression standardized by the beta-actin protein level. The ratio of cells without treatment was set at 1. (D) AN3CA (5 × 103 cells/well of 96-well plate) and HEC-59 (6 × 103 cells/well of 96-well plate) were treated with KJ-Pyr-9 (0, 2.5, 5, 10 and 20 μM), an inhibitor of c-Myc, for 72 h. Cell viability was analyzed by MTT assay. (E) Protein expressions of c-Myc, p53, KLK6, and survivin were determined by Western blot. ImageJ software was used to quantify the band intensities of c-Myc, p53, KLK6, and survivin. Data shown are the relative expression standardized by the beta-actin protein level. The ratio of cells without treatment was set at 1. N.D., not detected. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/34831139), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of c-Myc by Western Blot
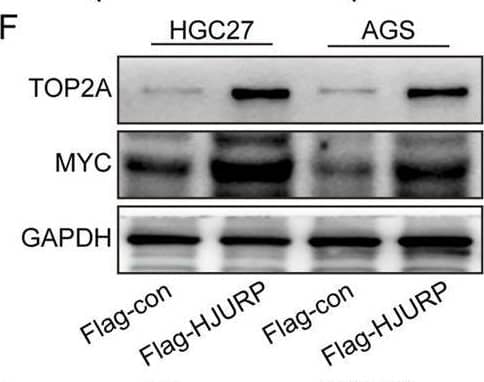
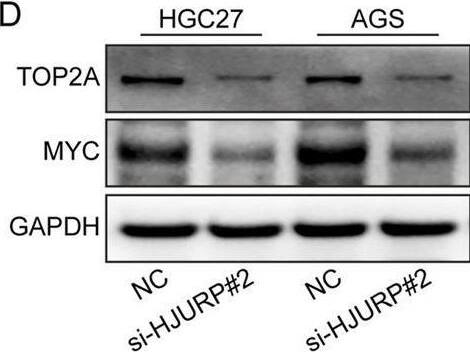
HJURP promotes proliferation and chemoresistance of GC cell by activating the TOP2A. (A) The relationship between the expression of TOP2A in Kaplan–Meier Plotter database and OS in GC patients. (B) The relationship between the expression of TOP2A in Kaplan–Meier Plotter database and FP in GC patients. (C, D) HGC27 and AGS were transfected with NC siRNA or HJURP siRNA for 48 h and then cell samples were collected, and the levels of TOP2A were determined by qRT-PCR and Western blot. (E, F) HGC27 and AGS were transfected with Flag-con or Flag-HJURP for 48 h and then cell samples were collected, and the levels of TOP2A were determined by qRT-PCR and Western blot. (G) CCK-8 assays were employed to evaluate the effect of HJURP knockdown with or without TOP2A overexpression on the proliferation of HGC27 GC cells. (H) Colony formation assays were employed to evaluate the effect of HJURP overexpression with or without TOP2A knockdown on the chemoresistance of HGC27 and AGS. **P < 0.01, ***P < 0.001, ****P < 0.0001. GC: gastric cancer, HJURP: Holliday Junction Recognition Protein, OS: overall survival, FP: first progression survival, DDP: cisplatin, TOP2A: Topoisomerase II alpha. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/40290723), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of c-Myc by Western Blot
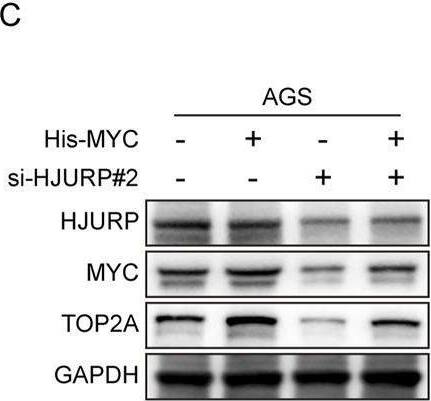
HJURP improves transcriptional activity of TOP2A via the promotion of MYC signaling. (A) The Venn diagram displays the potential transcription factors of TOP2A. (B, C) AGS were transfected with HJURP siRNA or His-MYC for 48 h and then cell samples were collected, and the levels of TOP2A, MYC and HJURP were determined by qRT-PCR and Western blot. (D–F) Genes are divided into 4 classes (Positive, Moderate, Weak, Negative) according to the expression level (visualized by contingency table heat map, and the depth of color represents the number of samples. The Pearson correlation of the two genes is calculated and Fisher’s exact test is performed. (G) The difference of MYC expression in TCGA-STAD dataset. (H) The ROC analysis of diagnostic accuracy for GC with MYC expression in TCGA databases. (I) The difference of TOP2A expression in TCGA-STAD dataset. (J) The ROC analysis of diagnostic accuracy for GC with TOP2A expression in TCGA databases. TCGA: The Cancer Genome Atlas, HJURP: Holliday Junction Recognition Protein, TOP2A: Topoisomerase II alpha. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/40290723), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of c-Myc by Western Blot
HJURP promotes proliferation and chemoresistance of GC cell by activating the TOP2A. (A) The relationship between the expression of TOP2A in Kaplan–Meier Plotter database and OS in GC patients. (B) The relationship between the expression of TOP2A in Kaplan–Meier Plotter database and FP in GC patients. (C, D) HGC27 and AGS were transfected with NC siRNA or HJURP siRNA for 48 h and then cell samples were collected, and the levels of TOP2A were determined by qRT-PCR and Western blot. (E, F) HGC27 and AGS were transfected with Flag-con or Flag-HJURP for 48 h and then cell samples were collected, and the levels of TOP2A were determined by qRT-PCR and Western blot. (G) CCK-8 assays were employed to evaluate the effect of HJURP knockdown with or without TOP2A overexpression on the proliferation of HGC27 GC cells. (H) Colony formation assays were employed to evaluate the effect of HJURP overexpression with or without TOP2A knockdown on the chemoresistance of HGC27 and AGS. **P < 0.01, ***P < 0.001, ****P < 0.0001. GC: gastric cancer, HJURP: Holliday Junction Recognition Protein, OS: overall survival, FP: first progression survival, DDP: cisplatin, TOP2A: Topoisomerase II alpha. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/40290723), licensed under a CC-BY license. Not internally tested by R&D Systems.