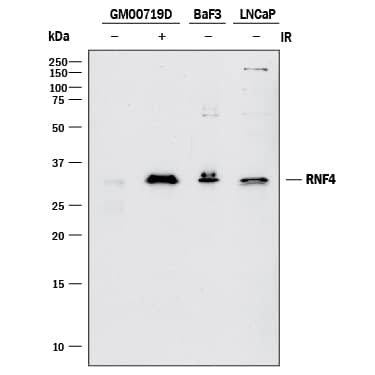
Detection of RNF4 by Western Blot
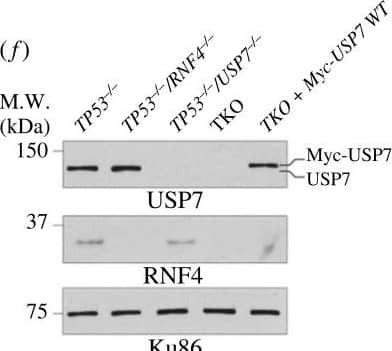
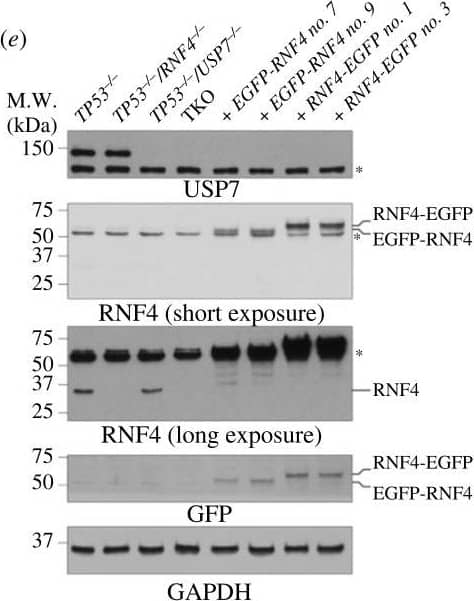
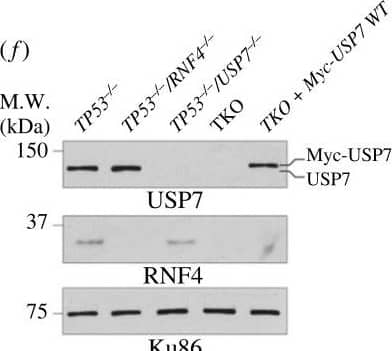
Bortezomib causes elevated cell apoptosis in TKO cells. (a) Representative dose–response curve to bortezomib measured by CellTiter-Glo cell viability assay. Cell survival is normalized to the untreated control. The mean survival with SD of each concentration is shown. The asymmetrical (five-parameter) logistic dose–response model is used to fit the curve. (b) Bortezomib IC50 values. Each data point is an IC50 value from an experiment. Bars and error bars indicate the mean with SD. (c) Cell apoptosis after 48 h of bortezomib measured by Annexin V-PI staining. Each data point is a biological replicate and the mean with SD is shown. (d) Cartoon schematics of constructs used to complement TKO cells. EGFP, enhanced green fluorescent protein; SIM, SUMO-interacting motif; RING, really interesting new gene; TRAF, tumour necrosis factor receptor-associated factor; CD, catalytic domain; UBL, ubiquitin-like. (e) Western blot analyses of whole cell extracts of TKO cells expressing either EGFP-RNF4 or RNF4-EGFP. (f) Western blot analyses of whole cell extracts of TKO cells expressing WT USP7. (g) Western blot analyses of SUMOylated chromatin-bound proteins of TKO cells expressing either EGFP-RNF4 or RNF4-EGFP. Cells were treated with 10 µM of MG132 and 1 mM of HU for 4 h before collecting. (h) Cell apoptosis after 48 h of bortezomib measured by Annexin V-PI staining in TKO and complemented cells. (b,c,h) Each data point represents the average value from independent plates in an experiment. Bars and error bars indicate the mean and SD across multiple biological experiments. Significance was measured using an ordinary one-way ANOVA with Tukey's multiple comparisons test; n.s., not significant; **p ≤ 0.01; ***p ≤ 0.001. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/37607592), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of RNF4 by Western Blot
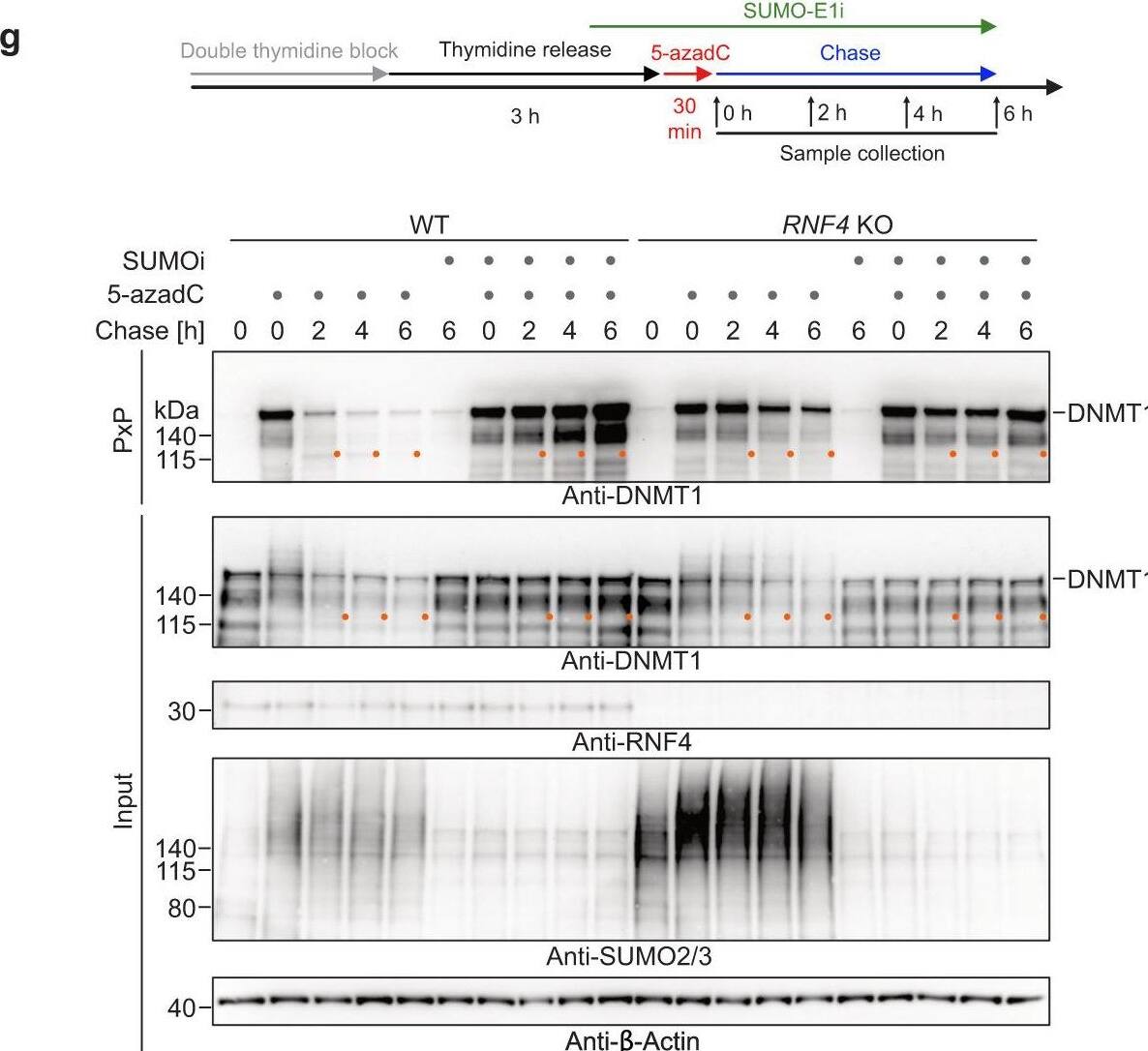
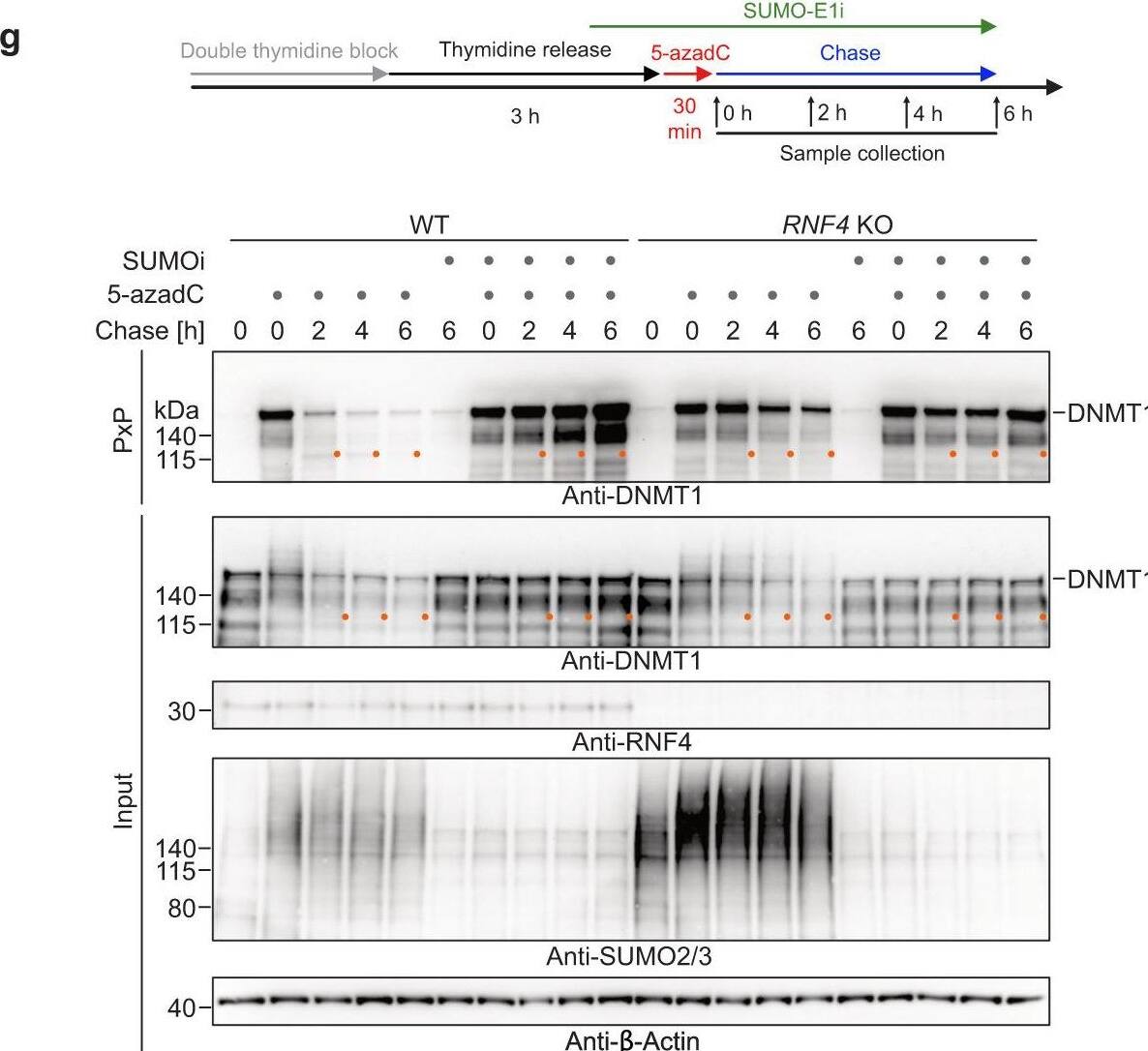
Global-genome repair of 5-azadC-induced DNMT1-DPCs monitored by PxP.a Schematic depiction of the experimental workflow used to monitor the repair of 5-azadC-induced DNMT1-DPCs. Cells were synchronized via a double thymidine block and released into early/mid S-phase for 3 h prior to induction of DNMT1-DPCs by a 30-min pulse of 5-azadC. Samples were collected either immediately after 5-azadC exposure or following a chase in drug-free media. Proteasome inhibitor (MG132, 5 µM), p97 inhibitor (p97i CB-5083, 5 µM) and SUMOylation inhibitor (SUMO-E1i ML-792, 5 µM) were added 1 h prior to induction of DPCs and kept during the chase with 5-azadC-free medium. Ubiquitylation inhibitor (Ub-E1i TAK-243, 1 µM) was added together with 5-azadC. b 5-azadC-induced DNMT1-DPC formation assessed by PxP. HeLa T-REx Flp-In cells were treated as depicted in (a) with the indicated doses of 5-azadC for 30 min prior to immediate isolation of DPCs using PxP and western blotting analysis. c–f 5-azadC-induced DNMT1-DPC formation and repair upon proteasome inhibition (c), inhibition of SUMOylation (d), inhibition of ubiquitylation (e), or knock-out of RNF4 (f) assessed by PxP. HeLa T-REx Flp-In cells were treated as depicted in (a) prior to extraction of DPCs using PxP, and analysis of samples by western blotting using the indicated antibodies. g HeLa WT and RNF4 knock-out (KO) cells were treated and analysed as depicted including an optional treatment with SUMOylation inhibitor (SUMO-E1i ML-792, 5 µM), prior to extraction of DPCs using PxP and analysis of samples by western blotting using the indicated antibodies. Experiments in (c–g) were repeated three times and similar results were obtained. Source data are provided as a Source Data file. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/36681662), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Human RNF4 by Western Blot
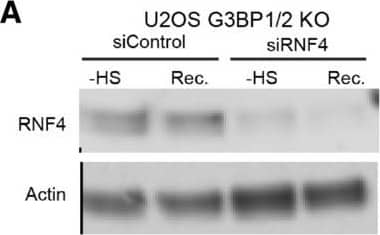
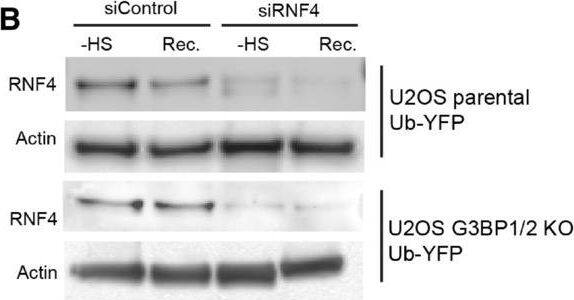
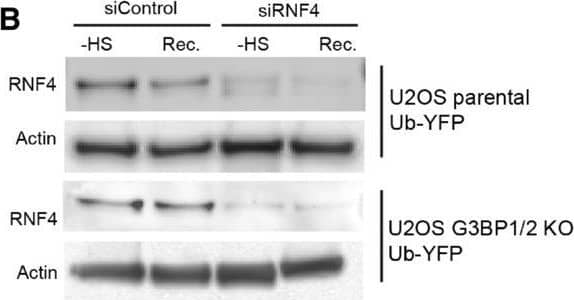
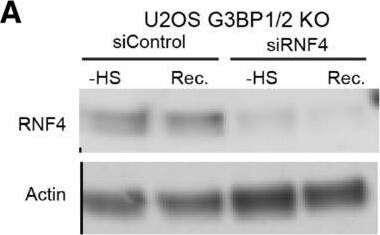
Inhibition of SUMO‐targeted ubiquitylation partially restores UPS activity in thermally stressed G3BP1/2 knockout U2OS G3BP1/2 knockout expressing Ub‐YFP were transfected with control siRNA or RNF4 siRNA. Cell lysates were analyzed by immunoblot with RNF4 & Actin antibodies. Parental U2OS & G3BP1/2 KO expressing Ub‐YFP were transfected with control siRNA or RNF4 siRNA. Cell lysates were analyzed by immunoblot with RNF4 & Actin antibodies. Fluorescence micrographs of parental U2OS stably expressing Ub‐YFP, which had been transfected with control or RNF4 siRNA. were either left untreated (− heat shock), or exposed to 43°C for 30 min & followed for 4 h after heat shock (Recovery). Scale bar is 10 μm.Fluorescence micrographs of U2OS G3BP1/2 KO stably expressing Ub‐YFP, which had been transfected with control or RNF4 siRNA. were either left untreated (− heat shock), or exposed to 43°C for 30 min & followed for 4 h after heat shock (Recovery). Scale bar is 10 μm.Immunoblotting for PML in control (siControl) & PML‐depleted (siPML) parental & G3BP1/2 KO U2OS . Quantification of mean cellular YFP fluorescence intensities in U2OS expressing Ub‐YFP with & without PML siRNA transfection. The YFP fluorescence is normalized to untreated control . The frequency & distribution of the relative fluorescence intensities per cell are shown as violin plots. The solid lines in each distribution represent the median, & dash lines represent the upper & lower interquartile range limits (n = 3 independent experiments, > 1,000 analyzed per condition, Kruskal‐Wallis test, *P < 0.05, **P < 0.01).Immunoblot of ataxin‐1 in parental & G3BP1/2 KO U2OS transfected with FLAGUb‐Ataxin‐1‐82Q.Immunoblot of FLAGUbiquitin in parental & G3BP1/2 KO U2OS transfected with FLAGUb‐Ataxin‐1‐82Q. Image collected & cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/36574355), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of RNF4 by Western Blot
Bortezomib causes elevated cell apoptosis in TKO cells. (a) Representative dose–response curve to bortezomib measured by CellTiter-Glo cell viability assay. Cell survival is normalized to the untreated control. The mean survival with SD of each concentration is shown. The asymmetrical (five-parameter) logistic dose–response model is used to fit the curve. (b) Bortezomib IC50 values. Each data point is an IC50 value from an experiment. Bars and error bars indicate the mean with SD. (c) Cell apoptosis after 48 h of bortezomib measured by Annexin V-PI staining. Each data point is a biological replicate and the mean with SD is shown. (d) Cartoon schematics of constructs used to complement TKO cells. EGFP, enhanced green fluorescent protein; SIM, SUMO-interacting motif; RING, really interesting new gene; TRAF, tumour necrosis factor receptor-associated factor; CD, catalytic domain; UBL, ubiquitin-like. (e) Western blot analyses of whole cell extracts of TKO cells expressing either EGFP-RNF4 or RNF4-EGFP. (f) Western blot analyses of whole cell extracts of TKO cells expressing WT USP7. (g) Western blot analyses of SUMOylated chromatin-bound proteins of TKO cells expressing either EGFP-RNF4 or RNF4-EGFP. Cells were treated with 10 µM of MG132 and 1 mM of HU for 4 h before collecting. (h) Cell apoptosis after 48 h of bortezomib measured by Annexin V-PI staining in TKO and complemented cells. (b,c,h) Each data point represents the average value from independent plates in an experiment. Bars and error bars indicate the mean and SD across multiple biological experiments. Significance was measured using an ordinary one-way ANOVA with Tukey's multiple comparisons test; n.s., not significant; **p ≤ 0.01; ***p ≤ 0.001. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/37607592), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of RNF4 by Western Blot
Bortezomib causes elevated cell apoptosis in TKO cells. (a) Representative dose–response curve to bortezomib measured by CellTiter-Glo cell viability assay. Cell survival is normalized to the untreated control. The mean survival with SD of each concentration is shown. The asymmetrical (five-parameter) logistic dose–response model is used to fit the curve. (b) Bortezomib IC50 values. Each data point is an IC50 value from an experiment. Bars and error bars indicate the mean with SD. (c) Cell apoptosis after 48 h of bortezomib measured by Annexin V-PI staining. Each data point is a biological replicate and the mean with SD is shown. (d) Cartoon schematics of constructs used to complement TKO cells. EGFP, enhanced green fluorescent protein; SIM, SUMO-interacting motif; RING, really interesting new gene; TRAF, tumour necrosis factor receptor-associated factor; CD, catalytic domain; UBL, ubiquitin-like. (e) Western blot analyses of whole cell extracts of TKO cells expressing either EGFP-RNF4 or RNF4-EGFP. (f) Western blot analyses of whole cell extracts of TKO cells expressing WT USP7. (g) Western blot analyses of SUMOylated chromatin-bound proteins of TKO cells expressing either EGFP-RNF4 or RNF4-EGFP. Cells were treated with 10 µM of MG132 and 1 mM of HU for 4 h before collecting. (h) Cell apoptosis after 48 h of bortezomib measured by Annexin V-PI staining in TKO and complemented cells. (b,c,h) Each data point represents the average value from independent plates in an experiment. Bars and error bars indicate the mean and SD across multiple biological experiments. Significance was measured using an ordinary one-way ANOVA with Tukey's multiple comparisons test; n.s., not significant; **p ≤ 0.01; ***p ≤ 0.001. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/37607592), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of RNF4 by Western Blot
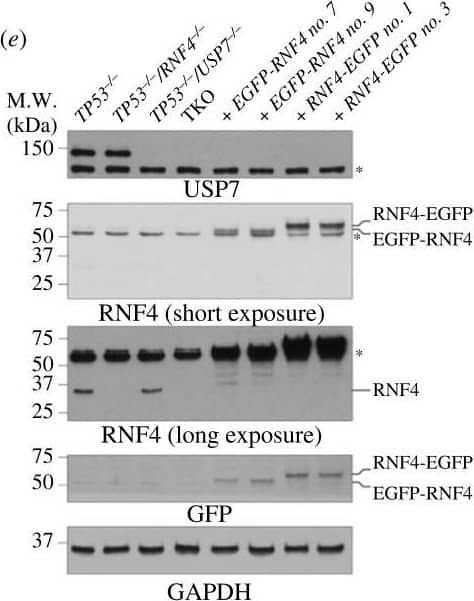
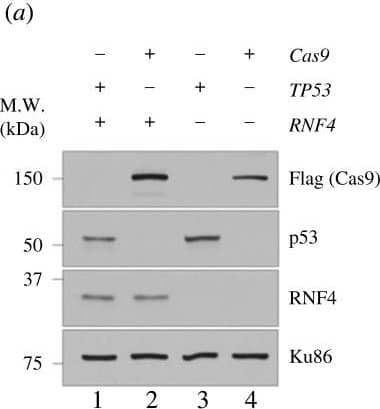
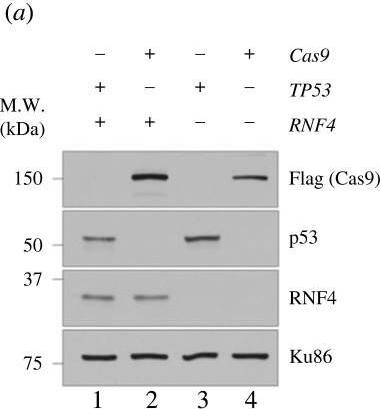
Genetic screens in TP53−/–RNF4-proficient and RNF4-deficient cells. (a) Western blot analyses of whole cell extracts from RPE-1 WT, RNF4−/– and TP53−/– mutants with Cas9 expression. Lanes 2 and 4 are samples isolated from the cell lines used in the genetic screens. (b) Flow chart of the CRISPR–Cas9 genetic screen in RPE-1 TP53−/– RNF4-proficient and RNF4-deficient cells using a custom, targeted DNA damage response sgRNA library. (c) Scatter plot showing the fitness of RNF4-proficient and -deficient cells between T0 and T18. The fitness is calculated as the log2 fold change of each gene between T0 and T18. Positive and negative genetic interactors are indicated in orange and blue, respectively. (d) Volcano plot showing the positive and negative genetic interactors of RNF4 at T18. The genetic interaction scores are the differential fitness between RNF4-proficient and -deficient cells. Positive and negative genetic interactors are indicated in orange and blue, respectively. Other genes of interest are shown in red. (e) Scatter plot showing the genetic interaction scores in two independent biological experiments for each gene. The Pearson correlation coefficient (R) and p-value are indicated. (f) Gene ontology (GO) analysis of genetic interactors (GIs) of RNF4 at T18. The top 10 positive (top) and negative (bottom) GO:BP terms and their z-scores are shown (see Material and methods). The significance (−log10p-value) of each term is indicated by the colour scale. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/37607592), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of RNF4 by Western Blot
Global-genome repair of 5-azadC-induced DNMT1-DPCs monitored by PxP.a Schematic depiction of the experimental workflow used to monitor the repair of 5-azadC-induced DNMT1-DPCs. Cells were synchronized via a double thymidine block and released into early/mid S-phase for 3 h prior to induction of DNMT1-DPCs by a 30-min pulse of 5-azadC. Samples were collected either immediately after 5-azadC exposure or following a chase in drug-free media. Proteasome inhibitor (MG132, 5 µM), p97 inhibitor (p97i CB-5083, 5 µM) and SUMOylation inhibitor (SUMO-E1i ML-792, 5 µM) were added 1 h prior to induction of DPCs and kept during the chase with 5-azadC-free medium. Ubiquitylation inhibitor (Ub-E1i TAK-243, 1 µM) was added together with 5-azadC. b 5-azadC-induced DNMT1-DPC formation assessed by PxP. HeLa T-REx Flp-In cells were treated as depicted in (a) with the indicated doses of 5-azadC for 30 min prior to immediate isolation of DPCs using PxP and western blotting analysis. c–f 5-azadC-induced DNMT1-DPC formation and repair upon proteasome inhibition (c), inhibition of SUMOylation (d), inhibition of ubiquitylation (e), or knock-out of RNF4 (f) assessed by PxP. HeLa T-REx Flp-In cells were treated as depicted in (a) prior to extraction of DPCs using PxP, and analysis of samples by western blotting using the indicated antibodies. g HeLa WT and RNF4 knock-out (KO) cells were treated and analysed as depicted including an optional treatment with SUMOylation inhibitor (SUMO-E1i ML-792, 5 µM), prior to extraction of DPCs using PxP and analysis of samples by western blotting using the indicated antibodies. Experiments in (c–g) were repeated three times and similar results were obtained. Source data are provided as a Source Data file. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/36681662), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of RNF4 by Western Blot
Genetic screens in TP53−/–RNF4-proficient and RNF4-deficient cells. (a) Western blot analyses of whole cell extracts from RPE-1 WT, RNF4−/– and TP53−/– mutants with Cas9 expression. Lanes 2 and 4 are samples isolated from the cell lines used in the genetic screens. (b) Flow chart of the CRISPR–Cas9 genetic screen in RPE-1 TP53−/– RNF4-proficient and RNF4-deficient cells using a custom, targeted DNA damage response sgRNA library. (c) Scatter plot showing the fitness of RNF4-proficient and -deficient cells between T0 and T18. The fitness is calculated as the log2 fold change of each gene between T0 and T18. Positive and negative genetic interactors are indicated in orange and blue, respectively. (d) Volcano plot showing the positive and negative genetic interactors of RNF4 at T18. The genetic interaction scores are the differential fitness between RNF4-proficient and -deficient cells. Positive and negative genetic interactors are indicated in orange and blue, respectively. Other genes of interest are shown in red. (e) Scatter plot showing the genetic interaction scores in two independent biological experiments for each gene. The Pearson correlation coefficient (R) and p-value are indicated. (f) Gene ontology (GO) analysis of genetic interactors (GIs) of RNF4 at T18. The top 10 positive (top) and negative (bottom) GO:BP terms and their z-scores are shown (see Material and methods). The significance (−log10p-value) of each term is indicated by the colour scale. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/37607592), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Human RNF4 by Western Blot
Inhibition of SUMO‐targeted ubiquitylation partially restores UPS activity in thermally stressed G3BP1/2 knockout U2OS G3BP1/2 knockout expressing Ub‐YFP were transfected with control siRNA or RNF4 siRNA. Cell lysates were analyzed by immunoblot with RNF4 & Actin antibodies. Parental U2OS & G3BP1/2 KO expressing Ub‐YFP were transfected with control siRNA or RNF4 siRNA. Cell lysates were analyzed by immunoblot with RNF4 & Actin antibodies. Fluorescence micrographs of parental U2OS stably expressing Ub‐YFP, which had been transfected with control or RNF4 siRNA. were either left untreated (− heat shock), or exposed to 43°C for 30 min & followed for 4 h after heat shock (Recovery). Scale bar is 10 μm.Fluorescence micrographs of U2OS G3BP1/2 KO stably expressing Ub‐YFP, which had been transfected with control or RNF4 siRNA. were either left untreated (− heat shock), or exposed to 43°C for 30 min & followed for 4 h after heat shock (Recovery). Scale bar is 10 μm.Immunoblotting for PML in control (siControl) & PML‐depleted (siPML) parental & G3BP1/2 KO U2OS . Quantification of mean cellular YFP fluorescence intensities in U2OS expressing Ub‐YFP with & without PML siRNA transfection. The YFP fluorescence is normalized to untreated control . The frequency & distribution of the relative fluorescence intensities per cell are shown as violin plots. The solid lines in each distribution represent the median, & dash lines represent the upper & lower interquartile range limits (n = 3 independent experiments, > 1,000 analyzed per condition, Kruskal‐Wallis test, *P < 0.05, **P < 0.01).Immunoblot of ataxin‐1 in parental & G3BP1/2 KO U2OS transfected with FLAGUb‐Ataxin‐1‐82Q.Immunoblot of FLAGUbiquitin in parental & G3BP1/2 KO U2OS transfected with FLAGUb‐Ataxin‐1‐82Q. Image collected & cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/36574355), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Human RNF4 by Western Blot
Inhibition of SUMO‐targeted ubiquitylation partially restores UPS activity in thermally stressed G3BP1/2 knockout U2OS G3BP1/2 knockout expressing Ub‐YFP were transfected with control siRNA or RNF4 siRNA. Cell lysates were analyzed by immunoblot with RNF4 & Actin antibodies. Parental U2OS & G3BP1/2 KO expressing Ub‐YFP were transfected with control siRNA or RNF4 siRNA. Cell lysates were analyzed by immunoblot with RNF4 & Actin antibodies. Fluorescence micrographs of parental U2OS stably expressing Ub‐YFP, which had been transfected with control or RNF4 siRNA. were either left untreated (− heat shock), or exposed to 43°C for 30 min & followed for 4 h after heat shock (Recovery). Scale bar is 10 μm.Fluorescence micrographs of U2OS G3BP1/2 KO stably expressing Ub‐YFP, which had been transfected with control or RNF4 siRNA. were either left untreated (− heat shock), or exposed to 43°C for 30 min & followed for 4 h after heat shock (Recovery). Scale bar is 10 μm.Immunoblotting for PML in control (siControl) & PML‐depleted (siPML) parental & G3BP1/2 KO U2OS . Quantification of mean cellular YFP fluorescence intensities in U2OS expressing Ub‐YFP with & without PML siRNA transfection. The YFP fluorescence is normalized to untreated control . The frequency & distribution of the relative fluorescence intensities per cell are shown as violin plots. The solid lines in each distribution represent the median, & dash lines represent the upper & lower interquartile range limits (n = 3 independent experiments, > 1,000 analyzed per condition, Kruskal‐Wallis test, *P < 0.05, **P < 0.01).Immunoblot of ataxin‐1 in parental & G3BP1/2 KO U2OS transfected with FLAGUb‐Ataxin‐1‐82Q.Immunoblot of FLAGUbiquitin in parental & G3BP1/2 KO U2OS transfected with FLAGUb‐Ataxin‐1‐82Q. Image collected & cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/36574355), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of RNF4 by Western Blot
Bortezomib causes elevated cell apoptosis in TKO cells. (a) Representative dose–response curve to bortezomib measured by CellTiter-Glo cell viability assay. Cell survival is normalized to the untreated control. The mean survival with SD of each concentration is shown. The asymmetrical (five-parameter) logistic dose–response model is used to fit the curve. (b) Bortezomib IC50 values. Each data point is an IC50 value from an experiment. Bars and error bars indicate the mean with SD. (c) Cell apoptosis after 48 h of bortezomib measured by Annexin V-PI staining. Each data point is a biological replicate and the mean with SD is shown. (d) Cartoon schematics of constructs used to complement TKO cells. EGFP, enhanced green fluorescent protein; SIM, SUMO-interacting motif; RING, really interesting new gene; TRAF, tumour necrosis factor receptor-associated factor; CD, catalytic domain; UBL, ubiquitin-like. (e) Western blot analyses of whole cell extracts of TKO cells expressing either EGFP-RNF4 or RNF4-EGFP. (f) Western blot analyses of whole cell extracts of TKO cells expressing WT USP7. (g) Western blot analyses of SUMOylated chromatin-bound proteins of TKO cells expressing either EGFP-RNF4 or RNF4-EGFP. Cells were treated with 10 µM of MG132 and 1 mM of HU for 4 h before collecting. (h) Cell apoptosis after 48 h of bortezomib measured by Annexin V-PI staining in TKO and complemented cells. (b,c,h) Each data point represents the average value from independent plates in an experiment. Bars and error bars indicate the mean and SD across multiple biological experiments. Significance was measured using an ordinary one-way ANOVA with Tukey's multiple comparisons test; n.s., not significant; **p ≤ 0.01; ***p ≤ 0.001. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/37607592), licensed under a CC-BY license. Not internally tested by R&D Systems.
Detection of Human RNF4 by Western Blot
Inhibition of SUMO‐targeted ubiquitylation partially restores UPS activity in thermally stressed G3BP1/2 knockout U2OS G3BP1/2 knockout expressing Ub‐YFP were transfected with control siRNA or RNF4 siRNA. Cell lysates were analyzed by immunoblot with RNF4 & Actin antibodies. Parental U2OS & G3BP1/2 KO expressing Ub‐YFP were transfected with control siRNA or RNF4 siRNA. Cell lysates were analyzed by immunoblot with RNF4 & Actin antibodies. Fluorescence micrographs of parental U2OS stably expressing Ub‐YFP, which had been transfected with control or RNF4 siRNA. were either left untreated (− heat shock), or exposed to 43°C for 30 min & followed for 4 h after heat shock (Recovery). Scale bar is 10 μm.Fluorescence micrographs of U2OS G3BP1/2 KO stably expressing Ub‐YFP, which had been transfected with control or RNF4 siRNA. were either left untreated (− heat shock), or exposed to 43°C for 30 min & followed for 4 h after heat shock (Recovery). Scale bar is 10 μm.Immunoblotting for PML in control (siControl) & PML‐depleted (siPML) parental & G3BP1/2 KO U2OS . Quantification of mean cellular YFP fluorescence intensities in U2OS expressing Ub‐YFP with & without PML siRNA transfection. The YFP fluorescence is normalized to untreated control . The frequency & distribution of the relative fluorescence intensities per cell are shown as violin plots. The solid lines in each distribution represent the median, & dash lines represent the upper & lower interquartile range limits (n = 3 independent experiments, > 1,000 analyzed per condition, Kruskal‐Wallis test, *P < 0.05, **P < 0.01).Immunoblot of ataxin‐1 in parental & G3BP1/2 KO U2OS transfected with FLAGUb‐Ataxin‐1‐82Q.Immunoblot of FLAGUbiquitin in parental & G3BP1/2 KO U2OS transfected with FLAGUb‐Ataxin‐1‐82Q. Image collected & cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/36574355), licensed under a CC-BY license. Not internally tested by R&D Systems.